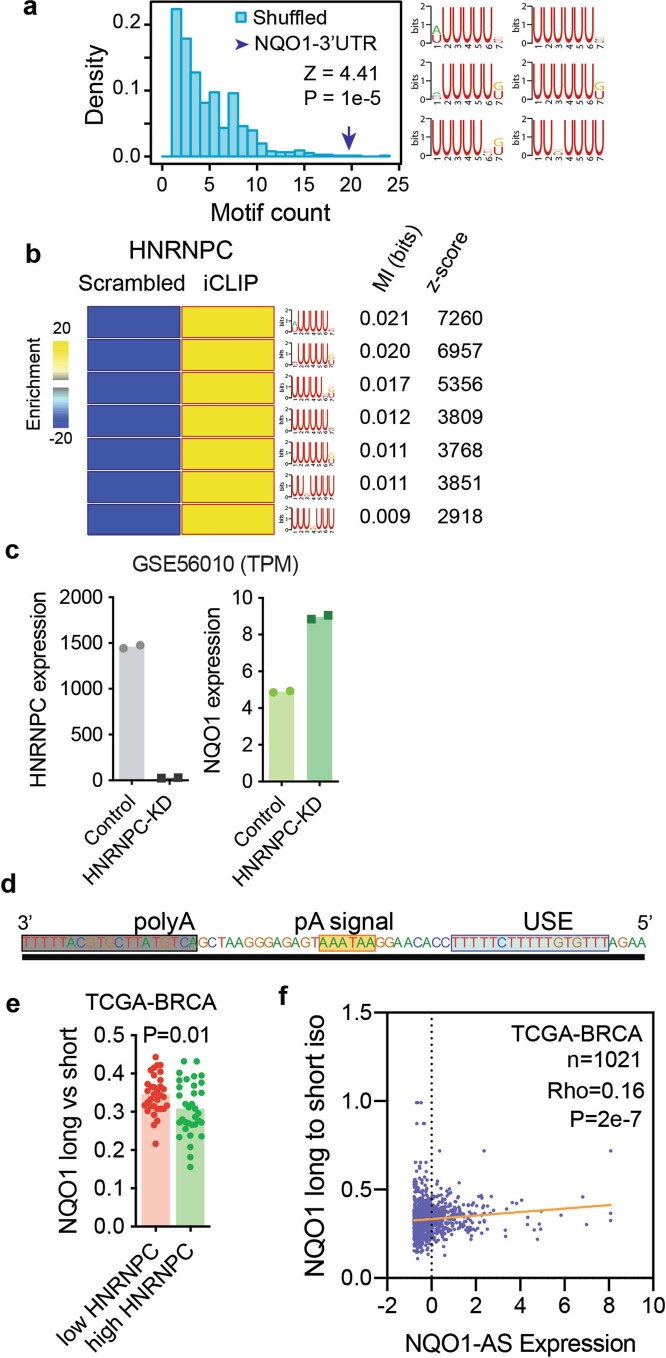
Extended Data Fig. 2. NQO1-AS binding masks HNRNPC binding sites which modulates polyA site selection.
(a) U-rich motif density in NQO1 3’UTR region complementary to NQO1-AS (purple arrow) relative to scrambled sequences with the same dinucleotide frequency (blue). (b) Heat map showing U-rich motif enrichment in HNRNPC iCLIP data relative to scrambled sequences. (c) Relative HNRNPC (left) and NQO1 (right) expression in HNRNPC knockdown and control (GSE56010; HEK293T) cells measured by RNA-seq. N = 2 independent cell cultures. (d) Region of NQO1 3’UTR highlighting two canonical polyadenylation sites. (e) Relative long vs short NQO1 isoform ratio in cells with high or low HNRNPC expression from TCGA-BRCA dataset (n = 33 and 24 patients respectively; ~top and bottom 5% of HNRNPC expression values). (f) Spearman correlation between NQO1 long to short isoform ratio and NQO1-AS expression in TCGA-BRCA dataset. N = 1021 patients. The P value in (a) was calculated using a shuffle test. The P value in (e) was calculated using a one-tailed Mann-Whitney U test.