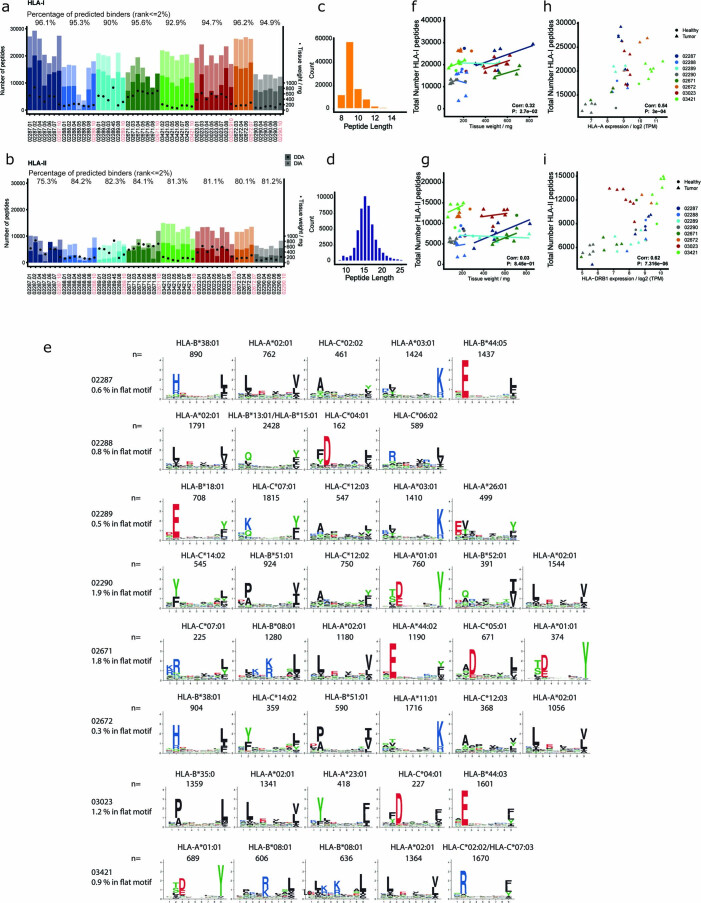
Extended Data Fig. 1. Mass spectrometry based immunopeptidomics performed on the different macro-region tissues.
a, HLA-I and b, HLA-II. DIA (light bars) analyses increased the number of identified peptides by up to 100% compared to DDA (solid bars). Peptide counts in the adjacent healthy macro-regions fall into the same range as the cancer samples. The average percentage of peptides predicted to bind the respective HLA allotypes in each patients are indicated above the bars. Peptide length distributions of c, HLA-I and d, HLA-II immunopeptidomics datasets. e Clustering of randomly selected 5000 HLA-I peptides per patient revealed the expected consensus binding motifs. Multiple specificity was observed for allele HLA-B*08:01 in patient 03421. The number of identified f, HLA-I (n = 102323 peptides) and g, HLA-II bound peptides (n = 53343 peptides) correlated with the starting tissue amount per patient (n = 53 macro-regions) but not across patients (p values 0.027 and 0.845, respectively). Across patients (N = 8 patients), a positive significant correlation was found between the number of identified h, HLA-I and i, HLA-II peptides with expression levels of HLA-A (p value 0.0003, Pearson cor= 0.54) and HLA-DRB1 (p value 7.3e-06, Pearson cor=0.62), respectively (n = 46 macro-regions with RNAseq and DIA data). For the correlation shown in f-i only macro-regions with DIA measurements were included (n = 53 macro-regions).