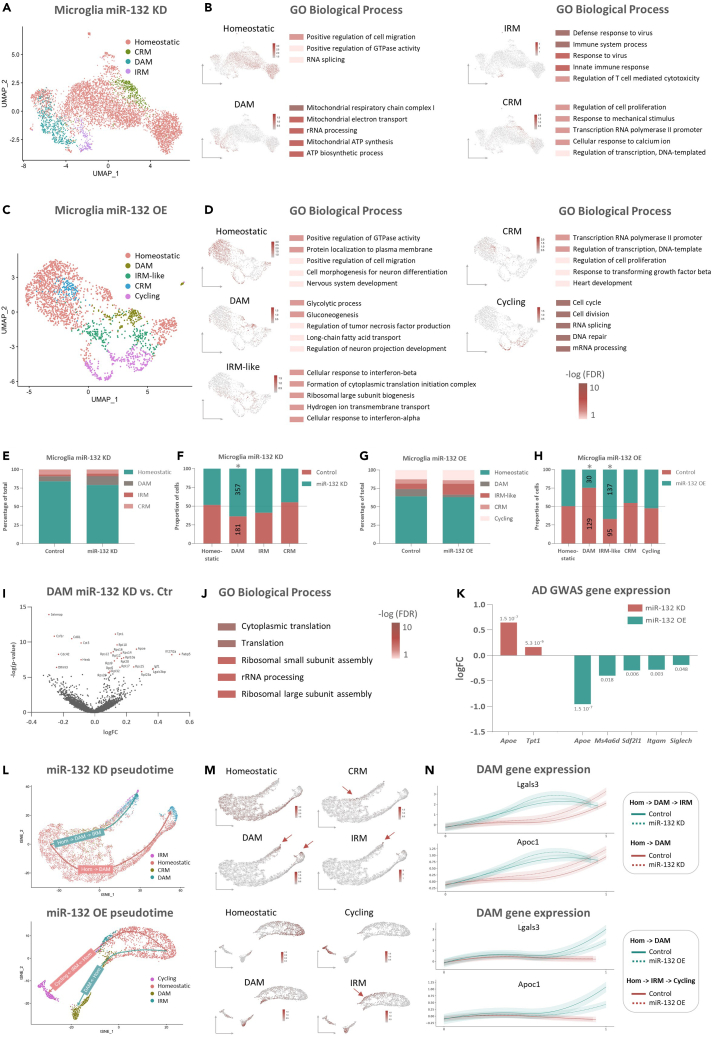
Figure 5.
miR-132 drives microglial cell state transitions
(A and C) Subset microglial population from miR-132 KD (A) or miR-132 OE (C) and corresponding control cells visualized on UMAP, colored according to identified clusters.
(B and D) UMAP plots colored based on the signature score of the combined gene set that characterizes each individual microglial subpopulation in miR-132 KD (B) or miR-132 OE (D). Top 5 unique GO biological processes of significantly enriched genes are provided per subtype; color is indicated according to significance (Fisher’s Exact test, p values corrected with FDR, adjusted p value <0.05 considered significant).
(E and G) Normalized percentage of cells present in distinct cellular states per condition in miR-132 KD (E) or miR-132 OE (G).
(F and H) Normalized percentage of cells present in distinct cellular states per cellular state in miR-132 KD (F) or miR-132 OE (H). Significance indicated with an asterisk (Chi-squared test, miR-132 KD DAM: p value = 0.0062; miR-132 OE DAM: p value = 0.0000004; miR-132 OE IRM: p value = 0.0007). The number of cells present in miR-132 KD/OE or control condition is depicted on the graph.
(I) Volcano plot showing DEGs comparing miR-132 KD cells to their corresponding controls in the DAM state. Significant DEGs indicated in red (Wilcoxon rank-sum test using Bonferroni for p value correction, adjusted p value <0.05 considered significant).
(J) GO biological processes significantly enriched in the top 500 most DEGs, with color indicating significance (Fisher’s Exact test, p values corrected with FDR, adjusted p value <0.05 considered significant).
(K) AD GWAS gene (adapted from153), p value cutoff set at 0.01) (Apoe, Tpt1, Ms4a6d, Sdf2l1, Itgam, Siglech) expression comparing miR-132 KD or OE cells with corresponding controls in the microglial population. Adjusted p values indicated (Wilcoxon rank-sum test using Bonferroni for p value correction, adjusted p value <0.05 considered significant).
(L) Single-cell trajectories of microglial cells obtained by pseudotime ordering using Palantir. Color according to identified clusters.
(M) UMAP plots colored based on the signature score of combined gene sets defining microglial cellular states. Arrows indicate populations of interest.
(N) Gene expression of the DAM-associated genes Lgals3 and Apoc1 along pseudotime. Color represents the pseudotime branch and type of line the experimental condition. Homeostatic/Hom, homeostatic microglia; DAM, disease-associated microglia; IRM, interferon-response microglia; CRM, cytokine-response microglia; Cycling, cycling microglia. See also Figure S3 and Table S4.