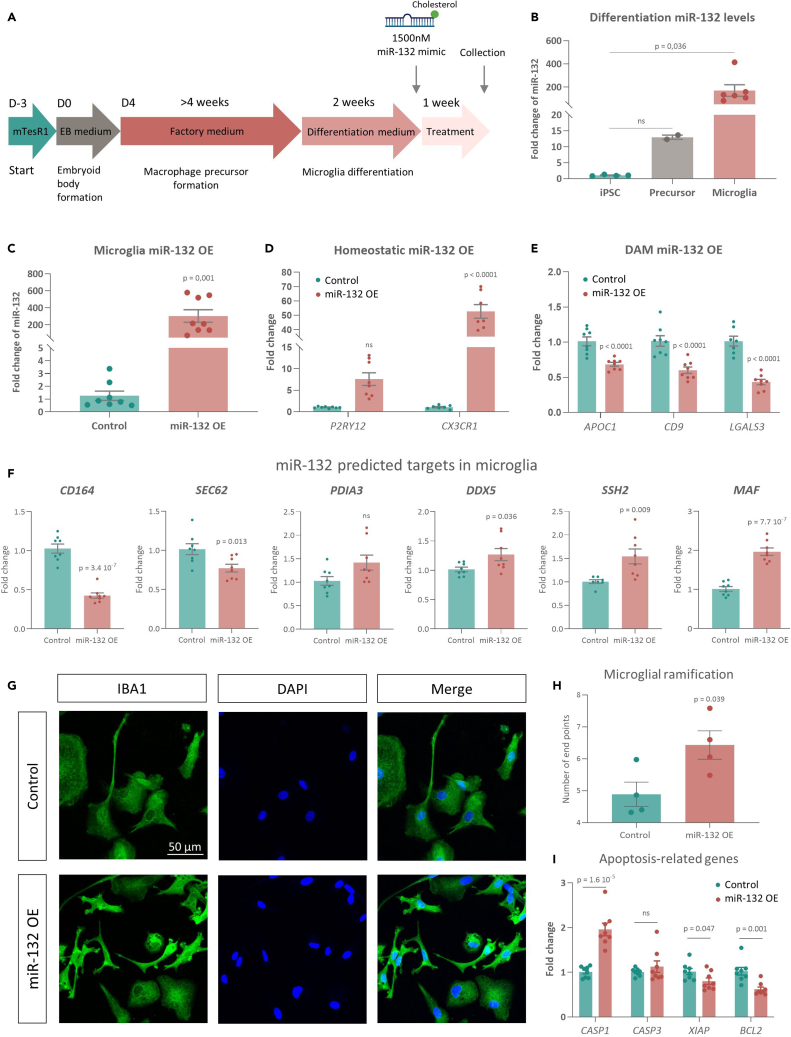
Figure 6.
Human iPSC-derived microglia acquire a homeostatic phenotype upon miR-132 supplementation
(A) Schematic representation of the differentiation protocol of human iPSCs into microglia. Cells were treated with cholesterol-conjugated miR-132 mimic or corresponding control oligonucleotide (1500 nM) for one week.
(B) Semi-quantitative real-time PCR of miR-132 levels along microglial differentiation. N = 2–6 biological replicates.
(C) Semi-quantitative real-time PCR of miR-132 levels in differentiated microglia after one week of treatment with a miR-132 mimic. N = 8 biological replicates.
(D and E) Semi-quantitative real-time PCR of marker genes characteristic of homeostatic microglia (P2RY12 and CX3CR1) (D) or DAM (APOC1, CD9 and LGALS3) (E) N = 7–8 biological replicates.
(F) Semi-quantitative real-time PCR of predicted miR-132 targets identified in the scRNAseq miR-132 OE experiment, as either downregulated in the microglial population (SSH2, PDIA3, DDX5, SEC62) or in the pseudo-bulked cells and also highly expressed in microglia (MAF and CD164).72 N = 8 biological replicates.
(G) Immunolabeling of IBA1 (green) and DAPI (blue) in miR-132- or control-treated iPSC-derived microglia. Merge indicates images obtained from overlaying images separately acquired with distinct channels.
(H) Morphological analysis of miR-132- or control-treated human iPSC-derived microglia, as quantified by the number of endpoints per cell. Each data point represents the average number of endpoints of 50 quantified cells, derived from two independent iPSC-to-microglia differentiations.
(I) Semi-quantitative real-time PCR of apoptosis-related genes comparing miR-132-treated human iPSC-derived microglia to controls. N = 8 biological replicates. Values are presented as mean ± SEM. In B, one-way ANOVA with Bonferroni correction was applied. In C, F and H, Student’s t test was used. In D, E and I two-way ANOVA with Tukey’s correction was used. See also Figure S1.