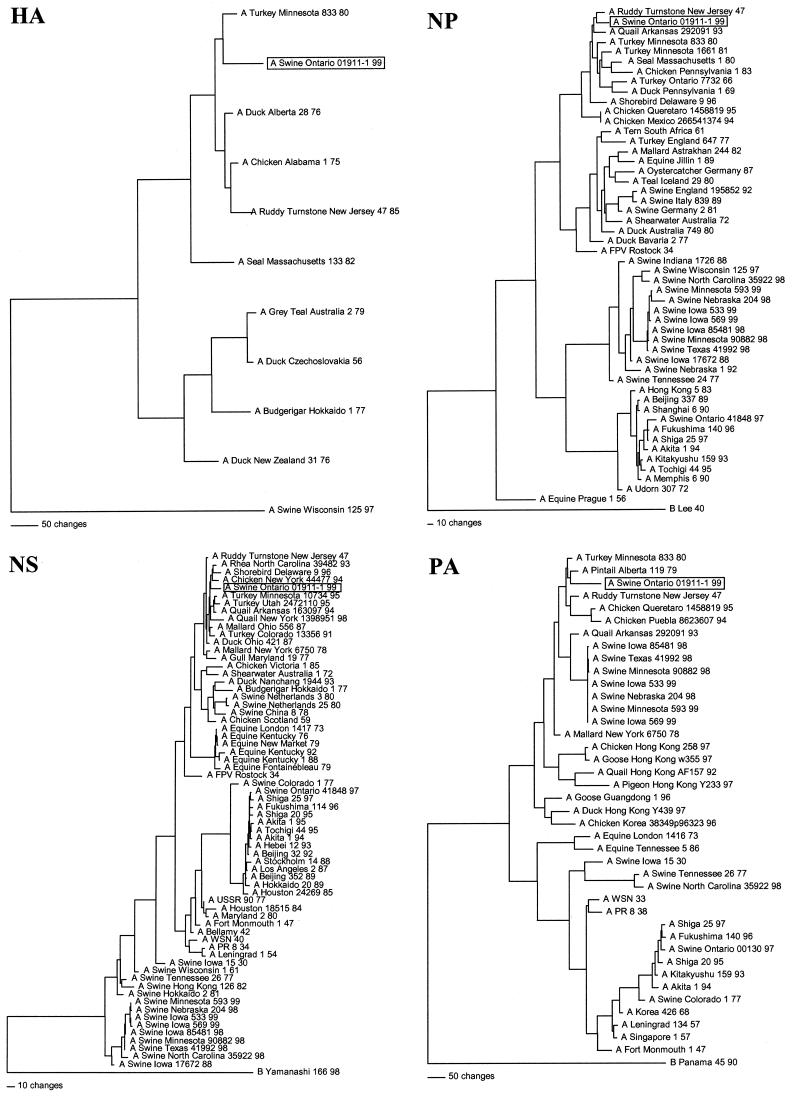
FIG. 1.
Nucleotide phylogenetic trees for the HA, NP, NS, and PA genes of Sw/ONT/99-1. The evolutionary relationships were estimated by the method of maximum parsimony (PAUP software, v.40b2; David Swofford, Smithsonian Institution, Washington, D.C.) by using the tree bisection-reconnection branch-swapping algorithm and with the MULTREES option and “gaps treated as missing” PAUP rule in effect. The trees shown represent the best of multiple rearrangements that were generated. The scores and number of rearrangements for each tree are as follows: HA, score = 1,911 of 684 rearrangements; NP, score = 2,460 of 48,866 rearrangements; NS, score = 1,313 of 2,013,155,259 rearrangements; and, PA, score = 3,286 of 24,377 rearrangements. The horizontal line distances are proportional to the minimum number of nucleotide changes needed to join nodes and gene sequences. The vertical lines are simply for spacing the branches and labels.