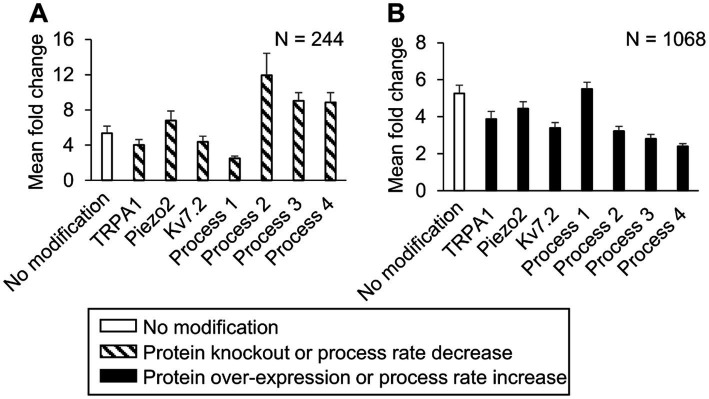
Figure 6.
In silico analysis identified ion channels and molecular processes that might contribute to inflammation-induced changes in the magnitude of action potential (AP) generation. We simulated either a knockout or a two-fold expression increase of three key model-identified ion channels (TRPA1, Piezo2, Kv7.1) and either an increase or a decrease in the rates of four key model-identified processes (GPCR phosphorylation, PKA inhibition, Gαq activation, and both Nav1.8 and Nav1.7 phosphorylation) using 10,000 randomly selected parameter sets. (A) Shown are the means and one standard error (SE) of the AP fold change from 244 simulations with seven modifications involving the individual knockout of proteins or 10-fold reduction of the rates of the four key processes (dashed bars). (B) Shown are the means and one SE of the AP fold change from 1,068 simulations with seven modifications involving a two-fold expression increase of the key proteins or a 10-fold increase in the rates of the four key processes (solid bars). In (A) and (B), the open bar indicates the mean and one SE of the magnitude of AP fold change in simulations with no modification. For implementing the modifications of process 4, we increased or decreased its rate by two-fold because the simulations with a 10-fold change did not run successfully. Process 1: G protein-coupled receptor phosphorylation, process 2: Gαq activation, process 3: protein kinase A inactivation, and process 4: Nav1.8 and Nav1.7 phosphorylation.