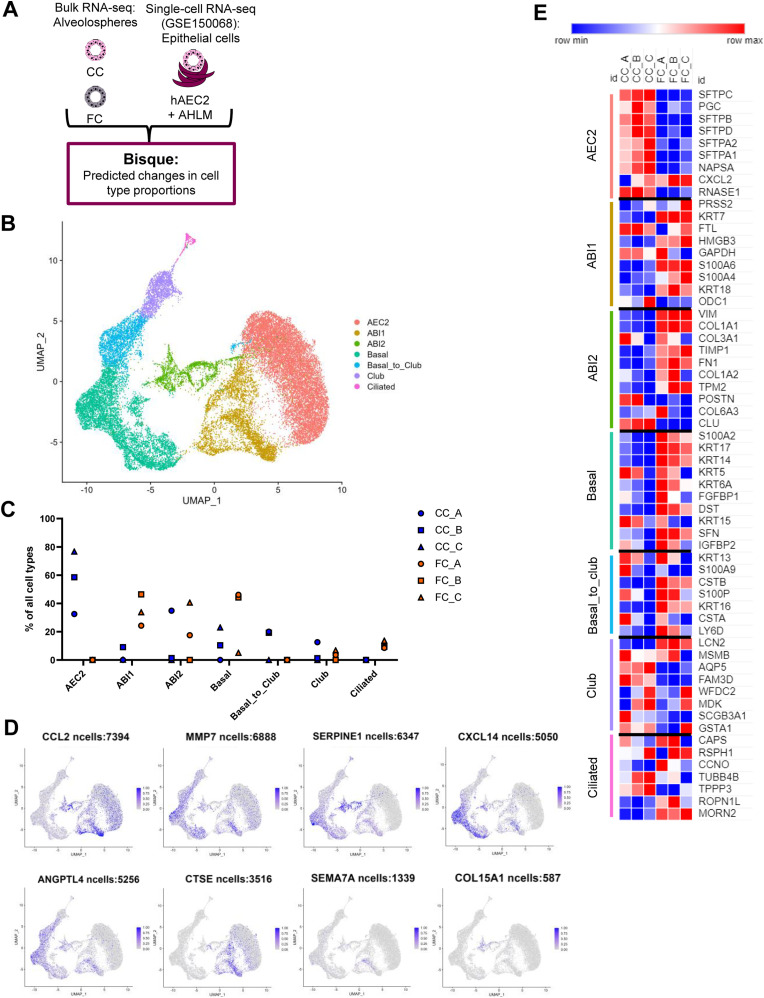
Figure 5. Deconvolution analysis predicts a trajectory of aberrant reprogramming through alveolar–basal intermediate states.
(A) Schematic depicting Bisque reference-based analysis of epithelial cell clusters from primary human AEC2 organoid co-cultures with adult human lung mesenchyme in the publicly available single-cell RNA sequencing dataset (GEO Accession: GSE150068). (B) UMAP plot of the identified cell clusters after re-annotating the deposited single-cell RNA-seq dataset GSE150068. (C) Cell proportions estimated by BisqueRNA in alveolospheres stimulated with CC or FC. n = 3 batches of alveolospheres. (D) Feature plots showing expression (scaled from 0 to 1) of selected up-regulated genes by FC stimulation. Genes were identified as described in the GitHub repository at: https://github.com/Lung-bioengineering-regeneration-lab/FC_alveolospheres. (E) Heatmap showing expression in alveolospheres of the most highly differentially expressed unique genes from each of the identified cell clusters. Genes selected by P(adj) < 0.05, absolute log2 fold change ≥ 0.7. See also Fig S9.