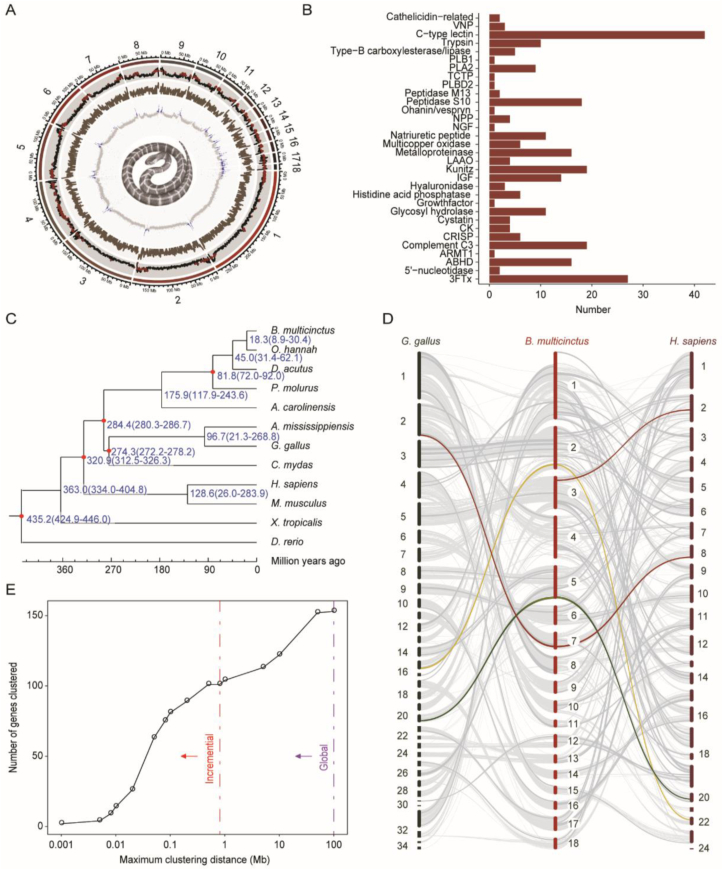
Figure 1.
The B. multicinctus genome indicated that toxin protein families originated independently from local tandem duplication. (A) The layers of the circus plot represent the chromosome length, repeat content, gene density and GC content (%) of the genome from the outside to the inside. Regions with repeat content greater than the third quantile are shown in brown and those with higher-than-average GC content are shown in blue. (B) Venom families identified in the genome. The identified proteins include members not expressed in the venom. Glycosyl hydrolase families (13,37,56) were presented as one. (C) Phylogenetic tree constructed using 387 single-copy orthologous genes conserved across 12 species speculates divergence times for each species. (D) Synteny maps of B. multicinctus aligned with H. sapiens and G. gallus. Light grey lines exhibit the synteny blocks. Dark grey lines exhibit the synteny blocks with venom genes. Red, yellow and green lines exhibit blocks with 3FTx, PLA2, and kunitz families respectively. (E) The incremental (red) and global (purple) tests show significant clustering of venom genes.