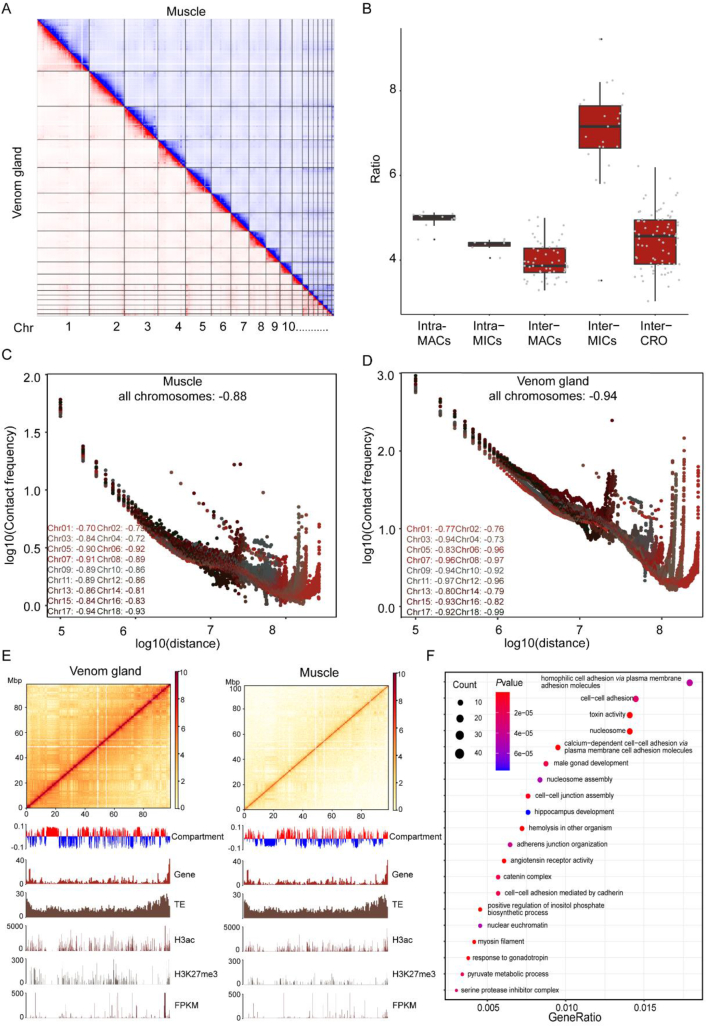
Figure 2.
3D chromatin organization and histone modification indicated the global epigenetic regulation of B. multicinctus genes. (A) Chromatin interaction in different tissues (red, venom gland; blue, muscle). (B) The increased ratio of chromosomal interactions in different chromosome groups when compare venom gland with muscle. (C, D) Whole-genome distance-contact frequency diagram with a 100-kb resolution in the venom gland (−0.94) and muscle (−0.88). Color: contact decay curve of different chromosomes; horizontal axis: distance between different sites on the chromosome; vertical axis: contact frequency. (E) Hi-C interaction map and colocalization of the genomic composition and various epigenetic marks on Chr7. The heatmap denotes the intrachromosomal Hi-C interaction frequencies among the pairwise 100-kb bins shown on the top. The PCA eigenvectors of the A and B compartments and genomic and epigenetic feature tracks are shown below in separate 100-kb bins, including abundances of genes and TEs, different histone modifications and mRNA expression levels (normalized by FPKM). (F) Bubble pattern of enriched pathways in the differential and conserved compartment domains (venom gland: A; muscle: B). The Y-axis represents the name of the pathway or function, and the X-axis represents the q values of each enriched gene family. The size of the bubble represents the number of genes in a signaling pathway or involved in a GO annotation. The color of the bubble indicates the ratio of the number of enriched genes.