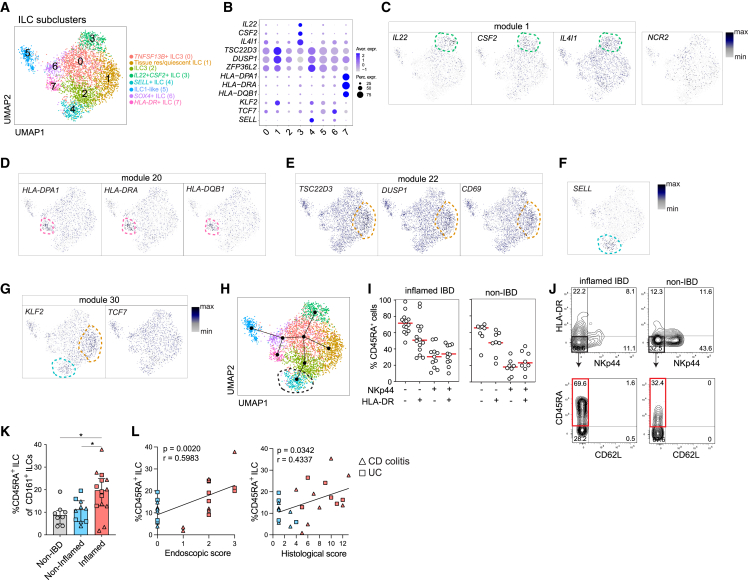
Figure 4.
Gene modules associate with ILC subclusters and identify quiescent/naive-like ILCs
(A) UMAP visualization of ILC subclusters after reclustering main cluster 1 shown in Figure 2B.
(B) Dotplot showing selected ILC-module genes (Figures 3D–3I). The expression is shown within the annotated subclusters of ILCs depicted in (A).
(C–G) UMAP visualization of selected ILC-module genes in the ILC subclusters. Dotted lines mark the subcluster where the depicted gene is highly and differentially expressed. A list of differentially expressed genes (DEGs) per ILC subcluster is shown in Figure S4A.
(H) Developmental trajectories of ILC subclusters performed with Monocle 3. Subcluster 4 (dotted circle) was used as the root.
(I) Frequencies of CD45RA+ ILCs among ILCs expressing combinations of HLA-DR and NKp44 in inflamed pIBD (n = 12–14) and non-IBD (n = 8) colon biopsies (bar graphs are shown as median). The samples were analyzed in independent experiments.
(J) Representative flow cytometry plots of CD45RA versus CD62L (marked in red is the CD45RA+ ILC subset), gated out of NKp44−HLA-DR− ILCs, in inflamed pIBD and non-IBD colon biopsies.
(K) Frequency of CD45RA+ ILCs out of CD161+ ILCs in non-ΙBD (n = 8), endoscopically non-inflamed (n = 10), and endoscopically inflamed pIBD (n = 14) colon biopsies. Mann-Whitney test (p < 0.05), mean with SD. The samples were analyzed in independent experiments.
(L) Correlation plots (Spearman, p < 0.05) between the frequency of CD45RA+ ILCs and the endoscopic and histological scores determined for each of the endoscopically non-inflamed (n = 8) and endoscopically inflamed (n = 14) pIBD colon samples.