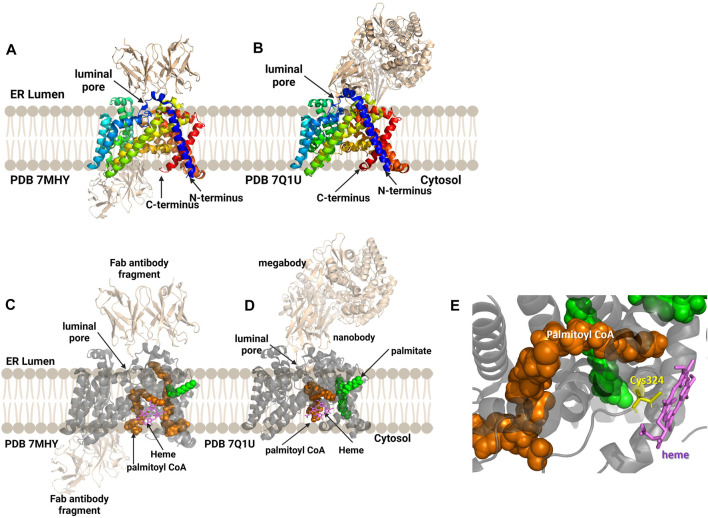
FIGURE 9.
Antibody-based complexes enabled cryoelectron microscopy determination of HHAT structure, revealing substrate binding sites, inhibitor binding, and an unanticipated heme cofactor. (A) Cryo-EM structure of HHAT by Jiang and co-workers (PDB ID 7MHY) shows the overall structure of the enzyme complexed to antibodies on both the cytoplasmic and luminal interfaces. (B) Binding to a designed megabody partner enabled the cryo-EM structure of HHAT by Coupland and co-workers (PDB ID 7Q1U). (C, D) Both groups report the presence of a heme (violet) in their structures; antibodies/megabody shown in beige, palmitoyl-CoA in orange, and palmitic acid in green. (E) Heme binding site in PDB ID 7Q1U showing heme iron coordination by Cys324. Figure created using Biorender.