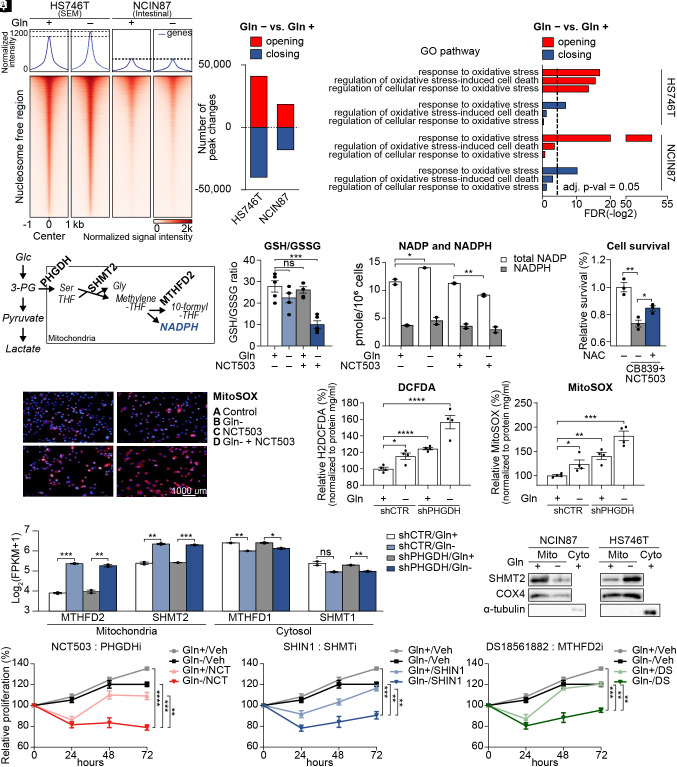
Fig. 2.
PHGDH-driven salvage pathway inhibits mitochondrial ROS production. (A) Chromatin accessibility heatmap of HS746T and NCIN87 with or without glutamine. Color intensity represents chromatin accessibility. (B) Numbers of peak changes after glutamine starvation were compared (adjusted P < 0.05). (C) Pathway analysis of the most variable peaks introduced in B associated with glutamine starvation. Dotted line indicates adjusted P-value = 0.05. (D) Diagram explaining the mitochondrial folate cycle using PHGDH, SHMT2, and MTHFD2 as a mediating enzymes. (E) GSH/GSSG ratio was measured with glutamine starvation and NCT503 treatment. (F) Total intracellular NADPH and NADP levels in glutamine starvation or NCT503-treated HS746T, normalized to cell number. (G) Cell survival with CB839 and NCT503 treatments in combination with N-acetyl-L-cysteine(NAC) (H) Mitochondrial superoxide visualized with MitoSOX with or without glutamine starvation and NCT503 treatment in HS746T cells. (I) Cellular ROS was measured via fluorescence assay using H2DCFDA in HS746T stable cell lines with or without glutamine. (J) Mitochondrial ROS was measured via fluorescence assay using MitoSOX in HS746T stable cell lines with or without glutamine. (K) RNA sequencing was performed on shControl or shPHGDH-stable HS746T cells with or without glutamine. Expression levels of mitochondrial folate cycle-related genes (MTHFD2 and SHMT2) and cytosolic folate cycle-related genes (MTHFD1and SHMT1) with or without glutamine were marked as log2(FPKM+1). (L) Immunoblot showing the mitochondrial protein level of SHMT2 upon glutamine starvation in HS746T and NCIN87 cells after mitochondrial isolation. (M–O) Proliferation assays for 72 h in HS746T cells. NCT503, PHGDH inhibitor; SHIN1, SHMT1 and SHMT2 inhibitor; DS18561882, MTHFD2 inhibitor. ns, no significance, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, Benjamini–Hochberg adjusted P-value for 2K and two-tailed Student’s t test for E–G, I, J, and M–O.