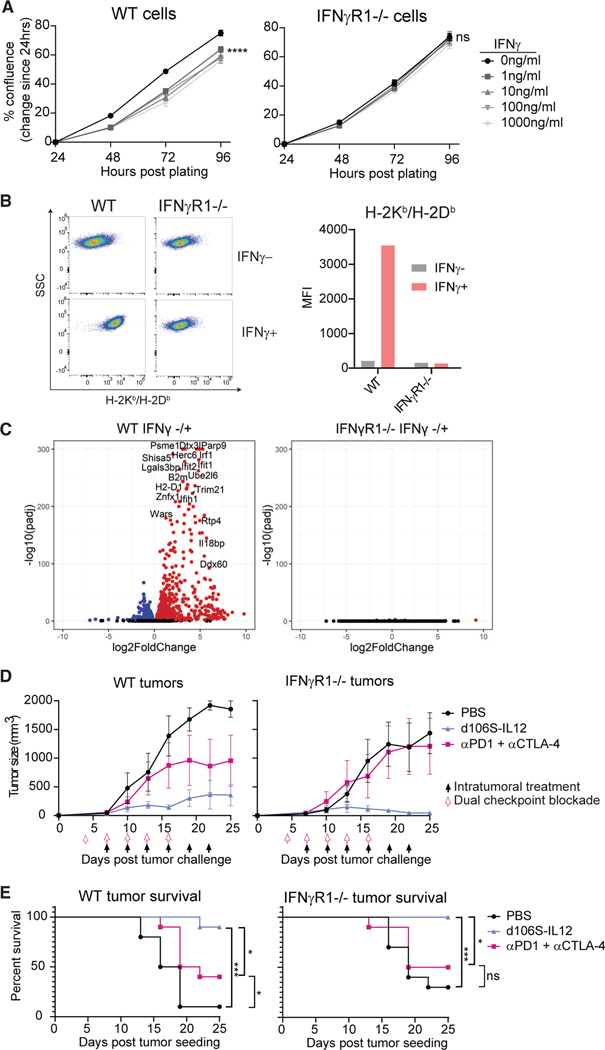
Figure 6. Tumors lacking IFNγ response are still controlled by d106S-IL12 therapy but not checkpoint blockade.
CRISPR-Cas9 gene editing was used to generate IFNγR1 (and STAT1; see also Figure S7) knockout B16Nectin1 cell lines.
(A) Wild-type or IFNγR1−/− cells were cultured with varying concentrations of IFNγ, and confluence was determined every 24 h by Celigo image cytometer.
(B) Wild-type or IFNγR1−/− cells were cultured in vitro with 50 ng/mL IFNγ and H2-Kb/Db (MHC-I) expression determined by flow cytometry 24 h later. Representative flow plots from multiple independent experiments.
(C) RNA sequencing was performed on wild-type or IFNγR1−/− cells cultured in vitro in the absence or presence (100 ng/mL) of IFNγ for 24 h (n = 3 per group). Differential gene expression was performed comparing IFNγ− versus IFNγ+ samples within the same genotype.
(D) Wild-type mice were challenged with either wildtype or IFNγR1−/− B16Nectin1 tumors and treated with PBS or d106S-IL12 starting at day 7 or anti-PD-1+ anti-CTLA-4 antibodies (150 mg each) i.p. starting at day 4 and proceeding every 3 days (N = 5 mice per group).
(E) Pooled survival across two experiments as in (D) (N = 10 mice per group).
Values are mean ± SEM, N = 3 unless otherwise noted. Confluence was compared by two-way ANOVA with Dunnett’s multiple comparisons test. Survival groups were compared using a log-rank test. *p < 0.05; ***p < 0.001; ****p < 0.0001.