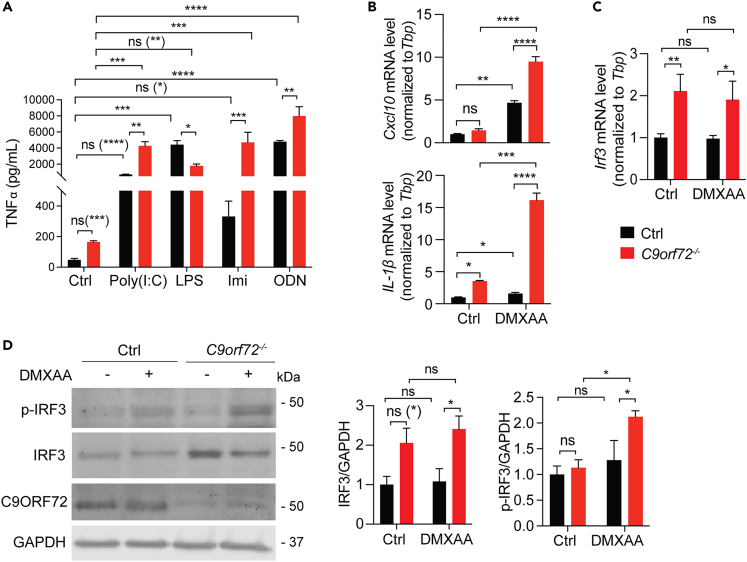
Figure 1.
Increased activation of the endosomal TLR and cGAS-STING signaling in C9orf72−/− cells
(A) ELISA analysis of TNFα levels in the cell medium of control and C9orf72−/− RAW264.7 cells treated 24h with TLR3 ligand poly(I:C) (1 μg/mL), TLR4 ligand LPS (1 μg/mL), TLR7 ligand imiquimod (Imi) (100 μM), or TLR9 ligand CpG ODN (1 μM). Data represent the mean ± SEM. Statistical significance was analyzed by two-way ANOVA (n = 3). Groups that exhibit non-significant differences with two-way ANOVA were re-analyzed using unpaired two-tailed Student’s t test, and the results are indicated in parentheses. ns = not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(B) RT-qPCR measurement of mRNA levels of cytokines Cxcl10 and Il1b in control and C9orf72−/− RAW264.7 cells without and with 16h of DMXAA (100 μg/mL) treatment. Data represent the mean ± SEM. Statistical significance was analyzed by two-way ANOVA (n = 3), ns = not significant, ∗p < 0.05, ∗∗∗∗p < 0.0001.
(C) RT-qPCR measurement of mRNA level of Irf3 in control and C9orf72−/− RAW264.7 cells before and after DMXAA treatment. Data represent the mean ± SEM. Statistical significance was analyzed by two-way ANOVA (n = 3). ns = not significant, ∗p < 0.05, ∗∗p < 0.01.
(D) Western blot analysis of IRF3 and p-IRF3 levels in control and C9orf72−/− RAW264.7 cells before and after DMXAA treatment. Data represent the mean ± SEM. Statistical significance was analyzed by two-way ANOVA (n = 3). Groups that exhibit non-significant differences with two-way ANOVA were re-analyzed using unpaired two-tailed Student’s t test, and the results are indicated in parentheses. ns = not significant, ∗p < 0.05.