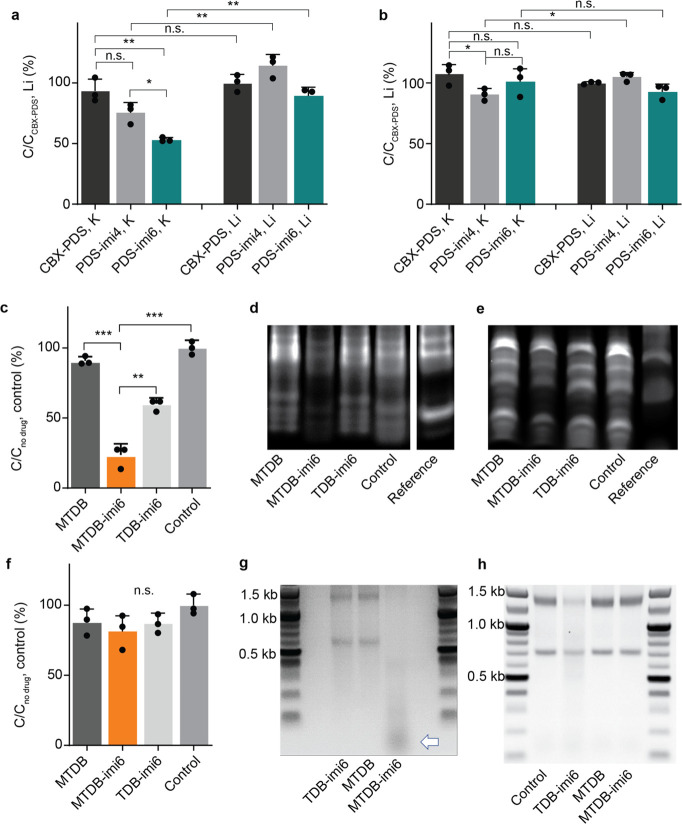
Figure 4.
PINADs degrade their target RNAs in vitro. (a) LC-MS validation of the G4 degraders against a G4-forming oligonucleotide after a 4 h 37 °C incubation with a specified molecule (n = 3). (b) LC-MS validation of the absence of cutting against a non-G4-forming oligonucleotide after a 4 h 37 °C incubation with a specified molecule (n = 3). (c) LC-MS validation of the pseudoknot degraders after a 3 h 37 °C incubation with a specified molecule (n = 3). (d) Nondenaturing gel validation of pseudoknot degraders on a 5′FAM-tagged pseudoknot oligonucleotide after a 6 h 37 °C incubation with a specified molecule. Double band for the pseudoknot might correspond to a pseudoknot monomer and dimer.24 (e) Denaturing gel validation of pseudoknot degraders on a 5′FAM-tagged pseudoknot oligonucleotide after a 6 h 37 °C incubation with a specified molecule. Representative gel shown (n = 3). (f) LC-MS validation of pseudoknot degraders losing efficiency when one of the pseudoknot stems is mutated and perturbs the pseudoknot secondary structure (n = 3). (g) Agarose gel electrophoresis validation of the cutting (white arrow) of extracted native SARS-CoV-2 RNA. (h) Agarose gel electrophoresis validation of no cutting of RNA extracted from HEK293FT cells. n.s. = not significant, *p < 0.05, **p < 0.01, ***p < 0.001.