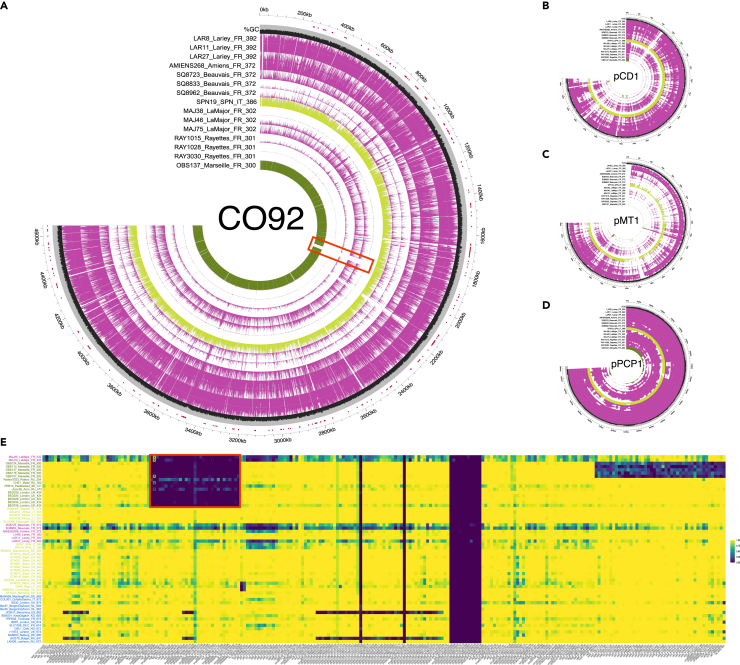
Figure 7.
Genome coverage and %GC variation
(A) CO92 Y. pestis reference genome.
(B) pCD1 plasmid.
(C) pMT1 plasmid.
(D) pPCP1 plasmid. A total of 13 samples sequenced in this study are shown (purple) together with two close relatives from the 17th c. CE (yellow) and the 18th c. CE (green) for comparison.
(E) Fraction of the gene covered at least once for 235 virulence and pathogenicity loci. Second pandemic Y. pestis genomes were considered as long as they were sequenced to a minimum 2-fold average depth-of-coverage. The red box highlights the 49-kb deletion identified, with “X” indicating those virulence and pathogenicity loci showing best Blast hits against environmental Yersinia species, and, thus, likely reflecting false positive alignments against Y. pestis. See also Figures S3, S4, and S7 and Table S3.