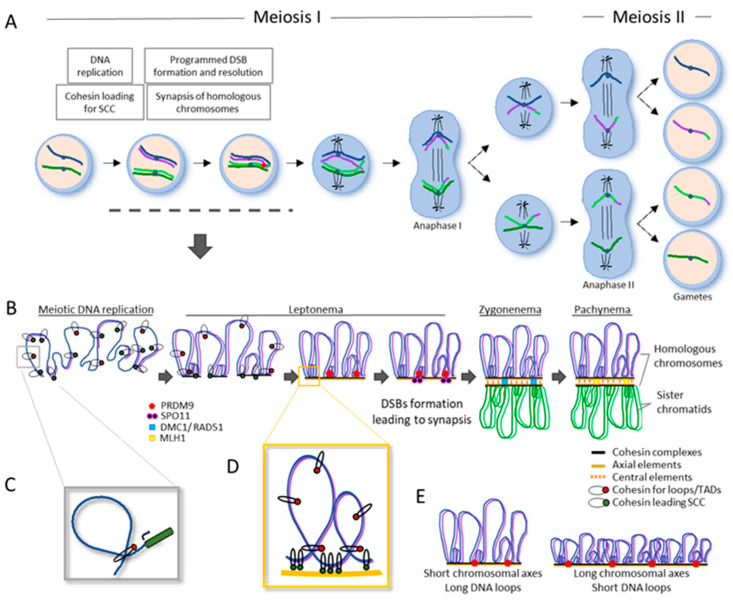
Figure 4.
Meiosis, key processes, and factors involved. (A) Different phases and key processes for Meiosis I and Meiosis II, from meiotic replication to the formation of haploid gametes. The use of only one pair of homologous chromosomes (blue and green) and their replication to sister chromatids (purple and light green) have been used for simplification. In Anaphase I, homologous chromosomes segregate, and in Anaphase II, sisters are pulled apart. Key processes during Prophase I (dashed line) are detailed in B, where different phases are represented. (B) Along meiotic DNA replication, cohesin complexes are loaded, leading to SCC (sister chromatid cohesion). From leptonema to pachytene, sister chromatids begin to condense along a proteinaceous structure formed by cohesin complexes, such as REC8, and axial elements, such as HORMAD1. Meiotic chromosomes are organized as DNA loops anchored to the axial elements, where proteins involved in the formation of DSBs, such as PRDM9 and SPO11, are recruited. These breaks are repaired where its resection leading to 3’ ends is key, covered by RPA and then by the recombinases RAD51 and DMC1 at zygonema. These two proteins catalyze strand invasion and will help with the homologous chromosomes’ recognition. In the meantime, that homologous recombination takes place, synapsis of homologous chromosomes occurs, with the polymerization of the central elements SYPs between both axials, leading to the Synaptonemal Complex (SC) formation. Using pachynema, SC is completed, and recombination intermediates can be resolved either as COs or non-COs. The resolution of COs is mediated by MLH1 proteins that will ultimately lead to chiasma formation. (C) cohesin occupancy mediates a role in transcription regulation by bringing two DNA regions together. (D) Schematic representation for distinct chromosomal localization pattern for cohesin mediating: SCC between sisters, loops and TADs formation, or transcription regulation. (E) The length of chromosomal axes and the size of DNA loops are inversely related, resulting in different numbers of DSBs formed by the protein SPO11. Figure adapted from [145].