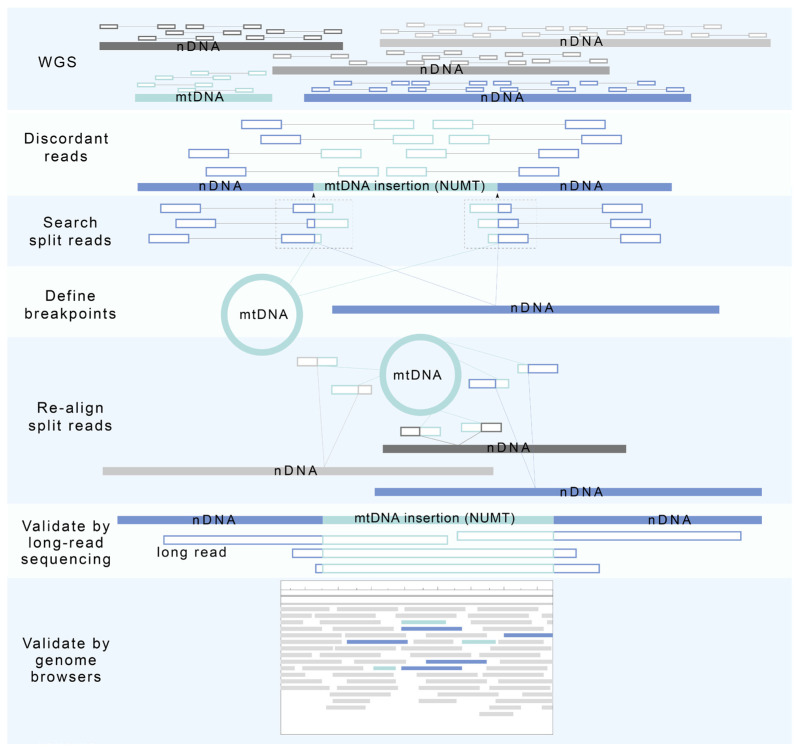
Figure 3.
A novel NUMT detection pipeline for paired-end WGS data. In this pipeline, discordant read pairs, with one read in the pair aligning to the nuclear DNA (nDNA) and the other read aligning to the mtDNA, are first detected and clustered to locate a putative breakpoint. With multiple split reads retained, a breakpoint can be identified. Split reads are then re-aligned to the reference mtDNA and nDNA to locate the origin of NUMT insertion. Long-read sequencing and genome browsers can be used to further validate the location of detected NUMTs.