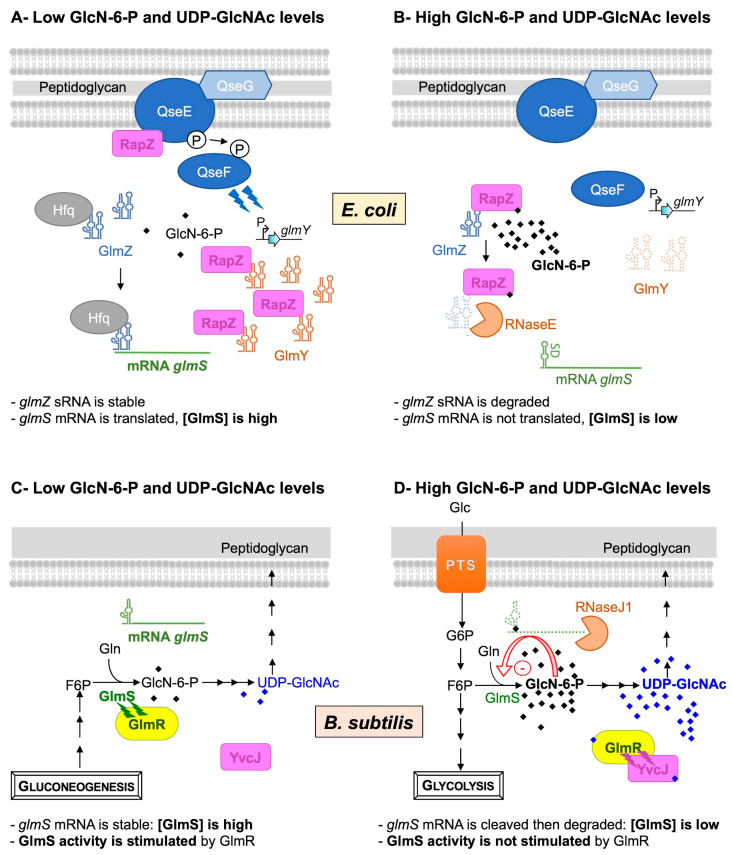
Figure 2.
Feedback regulation of GlmS in E. coli (A,B) and in B. subtilis (C,D). In E. coli, when the intracellular GlcN6P concentration is low (A), the two-component system QseE/QseF, associated with the lipoprotein QseG, boosts the expression of the sRNA GlmY that protects the second sRNA GlmZ from degradation and thus indirectly activates glmS. The sRNA GlmZ accumulates and interacts with the glmS mRNA through a base-pairing interaction, stabilized by Hfq, to stimulate its translation and increase the GlmS enzyme level. When the intracellular level of GlcN6P is high (B), RapZ binds to GlcN6P, thereby interfering with sRNA binding and leading to the stimulation of QseE/QseF. RapZ is released from complexes with GlmY, and the sRNA is rapidly degraded. Once free, RapZ binds and targets GlmZ sRNA to the RNase E endoribonuclease, which cleaves the sRNA at the base-pairing site, thus preventing the stimulation of glmS translation. In B. subtilis, when intracellular GlcN6P and UDP-GlcNAc concentrations are low (C), the ribozyme is not complexed to GlcN-6-P; the glmS transcript is stable and translated to increase the GlmS enzyme level. In addition, GlmR interacts with GlmS to stimulate its activity. When the intracellular GlcN6P and UDP-GlcNAc concentrations are high (D), GlcN6P binds to the ribozyme of the glmS transcript and stimulates its self-cleavage. No longer protected by a 5′ triphosphate end, the glmS transcript undergoes rapid exonucleolytic degradation by RNase J to decrease the GlmS enzyme level. In addition, GlmR binds UDP-GlcNAc and no longer interacts with GlmS to stimulate its activity. However, it binds to YvcJ, a protein homologous to RapZ.