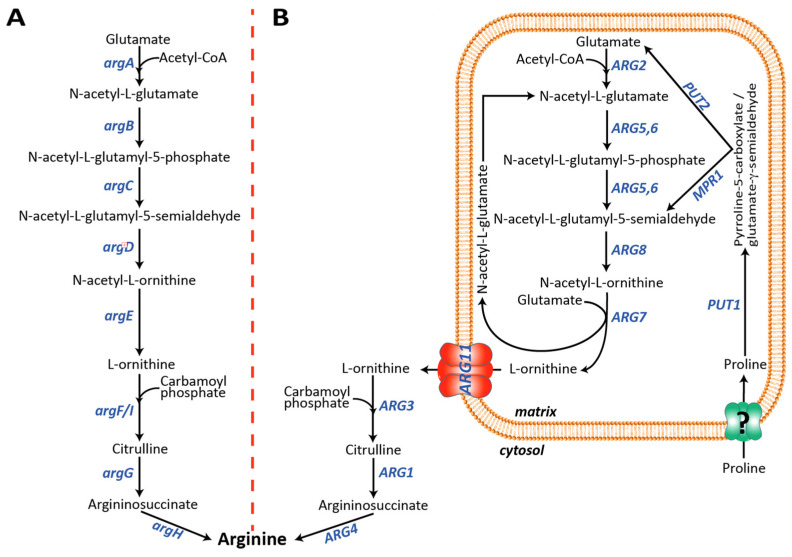
Figure 1.
Saccharomyces cerevisiae and Escherichia coli arginine biosynthetic pathways. (A) In E. coli, arginine biosynthesis occurs in the cytosol. argA, NAG synthase; argB, NAG kinase; argC, N-acetyl-glutamyl-5-phospate reductase; argD, N-acetyl-ornithine amino transferase; argE, N-acetyl ornithine deacetylase; argF/I, ornithine carbamoyl transferase; argG, argininosuccinate synthase; argH, argininosuccinate lyase. (B) In S. cerevisiae, the arginine biosynthesis starts in the mitochondrial matrix from the condensation of glutamate with acetyl-CoA catalyzed by ARG2 and ends with the synthesis of ornithine catalyzed by ARG7. Ornithine is then transported to the cytosol by ARG11, where it is finally converted to arginine. In S. cerevisiae the mitochondrial catabolism of proline may also produce N-acetyl-L-glutamyl-5-semialdehyde bypassing the first two steps of the main biosynthetic pathway. Although mechanistically the first three steps of the two pathways are not identical (see text for more details), the same metabolic intermediates are involved in both species. ARG2, NAG synthase; ARG5,6, N-acetylglutamate kinase and N-acetyl-5-glutamyl-phosphate reductase; ARG8, N-acetyl-ornithine aminotransferase; ARG7, N-acetyl-ornithine acetyltransferase; ARG11, mitochondrial ornithine transporter; ARG3, ornithine carbamoyl transferase; ARG1, argininosuccinate synthetase; ARG4, argininosuccinate lyase; PUT1, proline oxidase; PUT2, (S)-1-pyrroline-5-carboxylate dehydrogenase. MPR1, L-azetidine-2-carboxylic acid acetyltransferase.