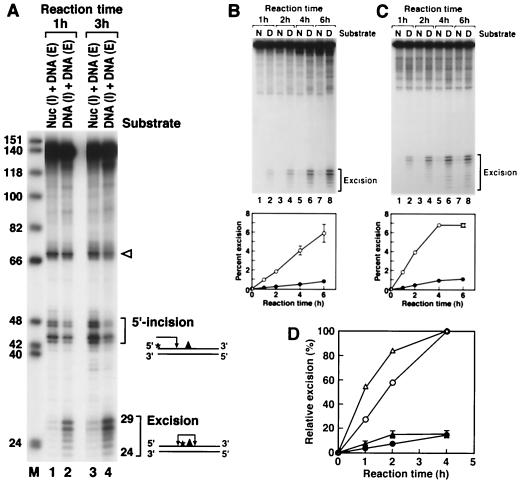
FIG. 3.
Effect of the nucleosome on nucleotide excision repair. (A) Inhibition of dual incision by the nucleosome tested by the incision assay and excision assay. Mixtures of end-labeled (E) or internally labeled (I) naked DNA (DNA) and internally labeled nucleosomal DNA (Nuc) were digested with the human excision nuclease, and the products were separated on an 8% denaturing polyacrylamide gel. The reaction mixtures contained 0.75 fmol each of end-labeled and internally labeled substrates. The same internally labeled DNA preparation was used either as naked DNA or in the form of nucleosome DNA in a mixture with end-labeled naked DNA. The sources of the reaction products are shown schematically at the right. Arrowhead, cleavage at the site of the (6-4) photoproduct resulting from excessive handling of the end-labeled DNA during substrate purification. The percentages of incision and excision in the various reactions were as follows. Lane 1, 0.9 (excision) and 5.4% (incision); lane 2, 5.5 (excision) and 4.0% (incision); lane 3, 2.3 (excision) and 11.6% (incision); lane 4, 12.8 (excision) and 6.2% (incision). (B) Kinetics of excision of (6-4) photoproducts from naked DNA and the nucleosome by reconstituted human excision repair nuclease. (Top) Reaction kinetics autoradiogram. Internally labeled (6-4) substrates in the form of nucleosomes (N) or naked DNA (D) were incubated with human excision nuclease for the indicated times, and the reaction products were analyzed on an 8% denaturing polyacrylamide gel. (Bottom) Kinetic plot of averages of three experiments including the one shown at the top. The percentage of the input substrate that was excised is plotted. Bars, standard deviations (those less than 0.07% are not shown). Open circles, naked DNA; solid circles, nucleosome. (C) Kinetics of excision of (6-4) photoproducts from naked DNA and the nucleosome by HeLa CE. (Top) Autoradiogram of a kinetics experiment. Internally labeled (6-4) substrates in the form of nucleosomes or naked DNA were incubated with HeLa CE for the indicated times, and reaction products were analyzed on an 8% denaturing polyacrylamide gel. (Bottom) Kinetics plot of averages of three experiments including the one at the top. The percentage of the input substrate that was excised is plotted. Standard deviations for all data points were less than 0.2%. Open circles, naked DNA; solid circles, nucleosome. (D) Kinetics of inhibition of (6-4) photoproduct excision by nucleosomes in HeLa (DDB+) and CHO (DDB−) CEs. Internally labeled (6-4) substrates in the form of nucleosomes or naked DNA were incubated in either HeLa CE or CHO AA8 CE, and the percent excision was determined as for Fig. 5. The values were expressed relative to the percent excision with naked DNA at the 4-h time point achieved by each CE and the averages of three experiments were plotted as relative excision. Circles, HeLa CE; triangles, CHO CE; open and solid symbols, naked DNA and nucleosomes, respectively. Bars, standard deviations. For HeLa CE, the data set shown in Fig. 3C was used.