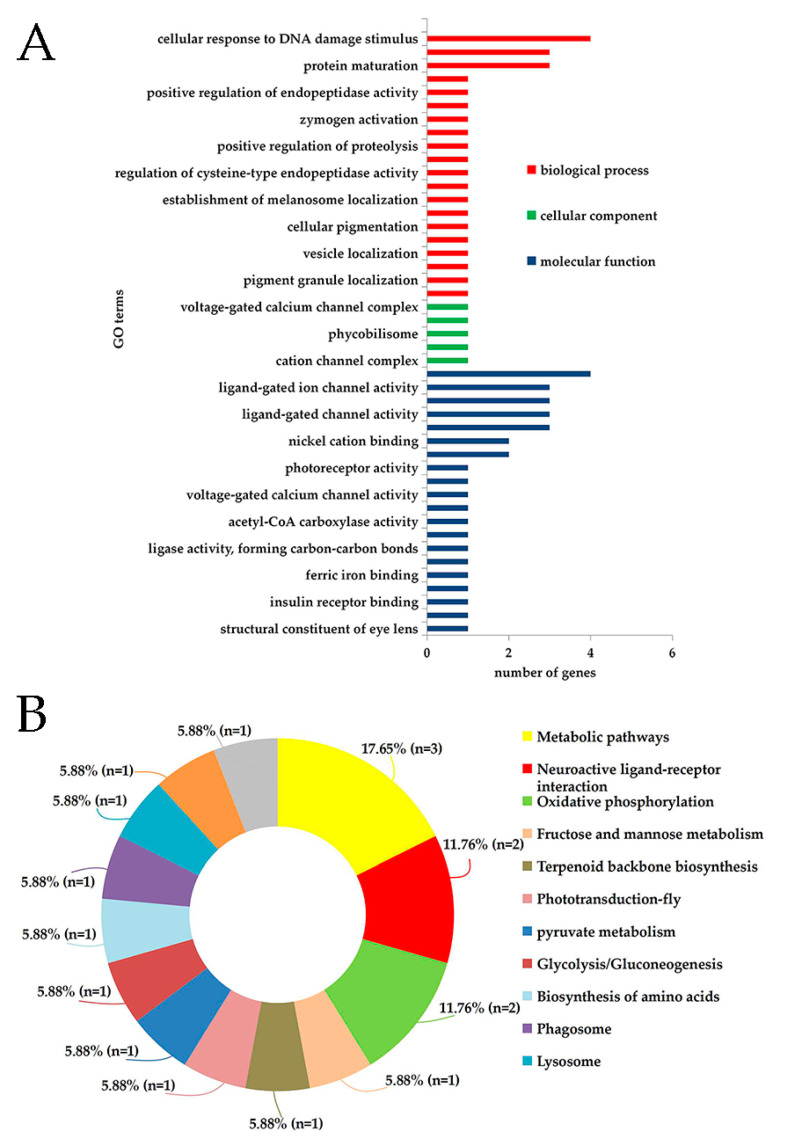
Figure 3.
Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) database of potential target genes of A. melliferra DE miRNAs annotation. (A) GO of potential target genes of the DE miRNAs of Apis melliferra. The DEmiRNAs were identified through the comparison of honey bees with strong olfactory performance with those with weak olfactory performance. The x-axis shows the GO term and the y-axis indicates the number of genes that were enriched in the three categories (biological process, cellular component, and molecular function). (B) KEGG database enrichment of potential target genes of the DE miRNAs of Apis melliferra were annotated to 13 pathways (Figure 3B). The scatter plot for KEGG enrichment results is shown in (Figure S2) and the mot enriched pathway terms is shown in (Table S1).