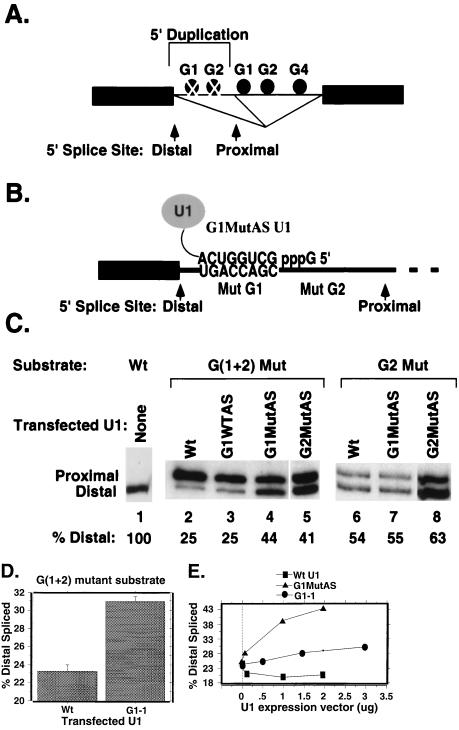
FIG. 7.
A complementary mutant U1 snRNA rescues 5′ splice site specificity when G triplets are mutated in a construct containing duplicated 5′ splice sites. (A) The 5′ splice site duplication substrates contained two identical 5′ splice sites separated by 58 nucleotides containing either wild-type (solid circles) or mutant (cross-hatched circles) G1 and G2 elements. The G1 and G2 mutations were identical to those indicated in Fig. 1A. The sequences downstream of the proximal 5′ splice site were wild type in each construct. Potential splicing pathways using either the distal or proximal 5′ splice sites are illustrated. (B) Model of potential base pairing between the G1MutAS U1 snRNA and a mutant (Mut) G1 element in the G(1+2) mutant duplication substrate. (C) RT-PCR (15-cycle) analysis of splicing products produced in transiently transfected HeLa cells. The positions of PCR products corresponding to RNAs spliced using the distal or proximal 5′ splice site are indicated. The substrates and cotransfected U1 genes used are indicated above the lanes. In the G(1+2) mutant, the G1 and G2 elements between the duplicated 5′ splice sites are both mutant. In the G2 mutant, the G1 element is wild type (Wt). The percentage of products resulting from usage of the distal site is indicated below each lane. (D) Rescue of distal 5′ splice site usage by expression of a U1 snRNA complementary to the first G triplet of the mutant G1 element (designated G1-1) in the G(1+2) mutant 5′ splice site duplication substrate. Means and standard errors were calculated from three experiments. (E) Graphical representation of distal splice site activation in the G(1+2) mutant substrate in HeLa cells cotransfected with increasing amounts of wild-type, G1MutAS, or G1-1 U1 expression plasmids.