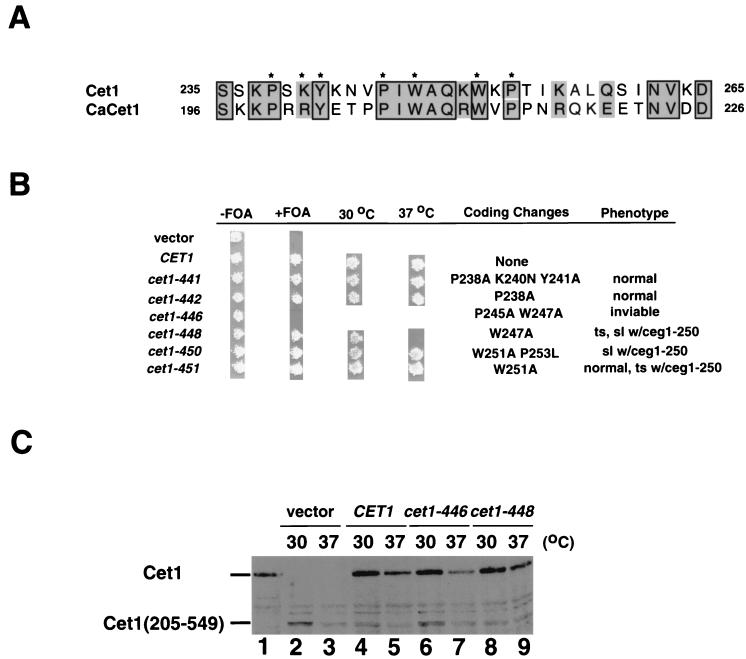
FIG. 2.
Analysis of the interaction region of Cet1. (A) Amino acid sequence alignment between Cet1 (GenBank accession no. AB008799, residues 235 to 265 [39]) and CaCET1 (AB016242, residues 196 to 226 [44]). Asterisks indicate the residues of Cet1 mutated in this study. (B) Phenotypes of site-directed mutants. Wild-type CET1 and six mutant alleles were transformed into YSB533 (−FOA) and tested for complementation by plasmid shuffling (+FOA). Alleles that supported viability were further tested for growth at 30 and 37°C. The coding changes and phenotypes of the alleles are listed to the right. ts, temperature sensitivity; sl, synthetic lethality. ceg1–250 is a temperature-sensitive allele of CEG1 (7). (C) Immunoblot analysis of mutant proteins. YSB710 containing the Cet1(205–549) allele was transformed with pRS313 (lanes 2 and 3), pRS313-CET1 (lanes 4 and 5), pRS313-cet1-446 (lanes 6 and 7), and pRS313-cet1-448 (lanes 8 and 9). Lane 1 had the extract from wild-type yeast. Cultures of 100 ml of the indicated strains were grown at 30°C to an optical density at 600 nm of 0.4. Cultures were split, resuspended in 50 ml of medium prewarmed to the indicated temperature, and further cultured for 90 min at that temperature. Twenty micrograms of whole-cell extract from each culture was analyzed by immunoblotting with anti-Cet1 antibody.