Figure 3.
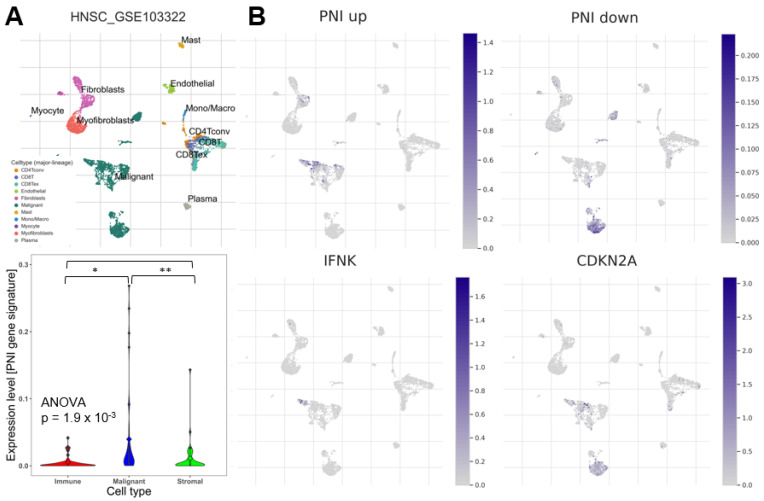
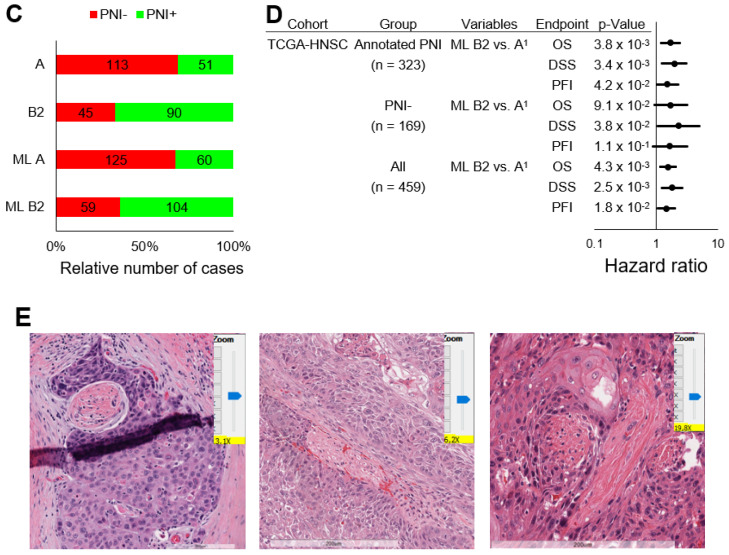
Prominent expression of the PNI-related 44-gene signature in cancer cells and establishment of a PNI classifier by a random forest model. (A) UMAP plot visualizing the different cell types given in GSE103322 [17], with included figure legend (malignant, endothelial, mast cells, plasma, myocyte, myofibroblasts, fibroblasts, CD4, CD8, monocytes, macrophages). Violin plot demonstrating the statistical significance of malignant cell scores (blue) compared to immune (red) and stromal (green) cell scores based on the 44-gene set. * p < 0.05, ** p < 0.005 as determined by an ANOVA and Tukey’s post hoc test. (B) UMAP plots for the 37 downregulated (PNI down) and 7 upregulated (PNI up) DEGs of the 44-gene signature as well as CDKN2A and IFNK illustrating the expression of these genes in distinct cell types in GSE103322 [17]. (C) The bar plot summarizes the relative frequency of PNI− (red) or PNI+ (green) tumors from TCGA-HNSC with an annotated PNI status, which were classified as either cluster A and subcluster B2 by the PNI-related 44-gene signature or ML A or ML B by the ML model. (D) Forest plot for 5-year OS, DSS, and PFI based on univariate Cox regression models for patients from the indicated groups of TCGA-HNSC, which were stratified by the ML model into ML A or B. 1 Reference group. (E) Representative pictures of digital images for H&E-stained slides from the Cancer Digital Slide Archive of TCGA-HNSC confirms the presence of PNI for clinically PNI-annotated tumors and tumors without PNI annotation, which were predicted as ML B2 by the random forest model.