Figure 1.
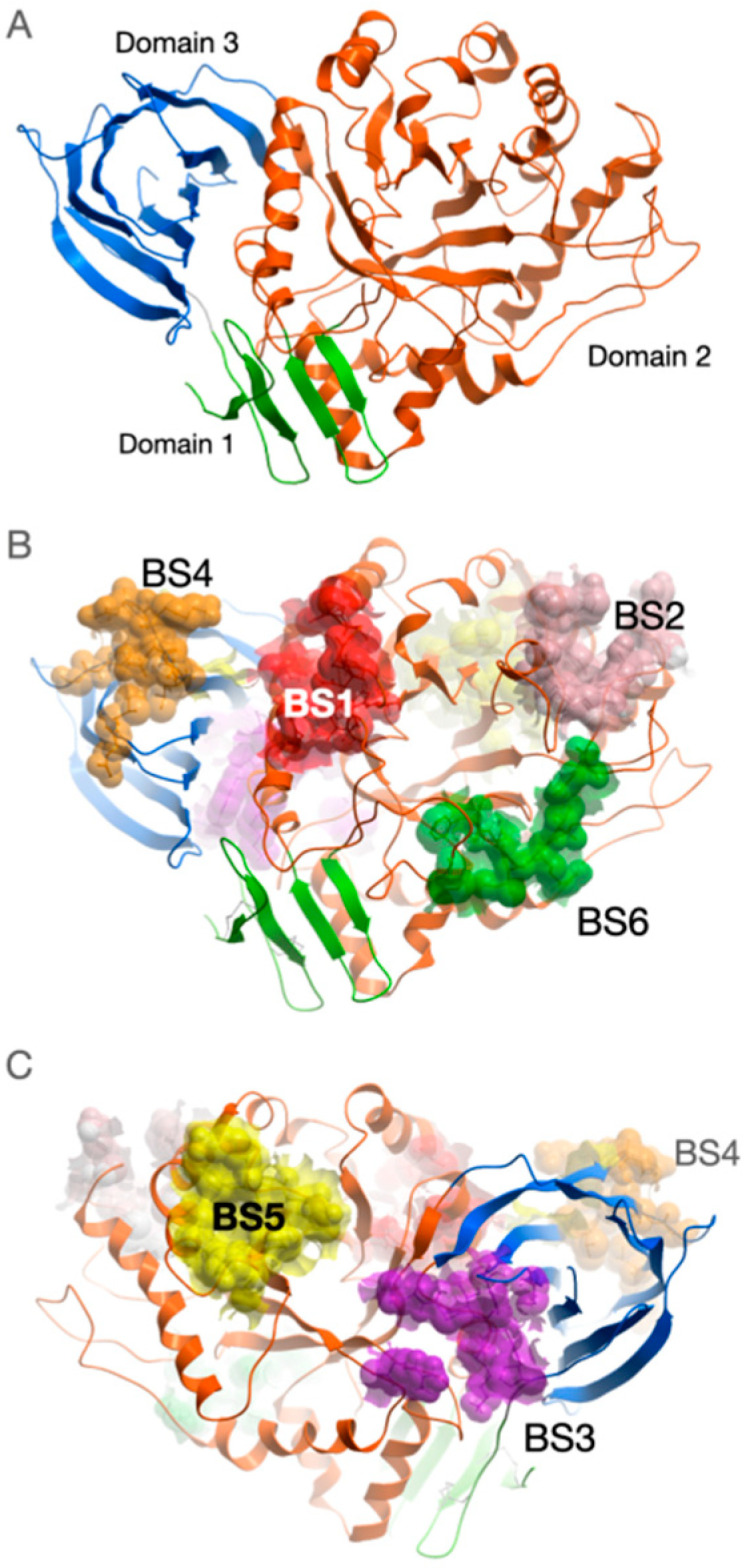
Spatial structure of GCase with potential binding sites mapped on its surface. (A) GCase comprises three domains marked by different colors: green—Domain 1, orange—Domain 2, blue—Domain 3. Amino acid residues combined into potential binding sites (BS) are displayed by van der Waals spheres: (B) view from the side of GCase where the active site is located (red—BS1, pink—BS2, green—BS6, orange—BS4), (C) view from the opposite side of protein (violet—BS3, orange—BS4, yellow—BS5). The model of the mutant form of the enzyme was constructed by introducing the amino acid substitution N370S into the structure deposited in 2NT1 PDB entry (subunit A) followed by relaxation using molecular dynamics simulations in explicit water.
