Table 2. Exemplar SOLES project – AD-SOLES workflow.
| Stage | Details | Integrated tools |
|---|---|---|
| Systematic search | ● Searches conducted using general Alzheimer’s disease search terms across Web of Science, Scopus, EMBASE and PubMed ● Customised R packages which make use of APIs to retrieve data on a weekly basis for three databases, while EMBASE is searched manually every 6 months ● Additional metadata for each study retrieved from CrossRef and OpenAlex |
rcrossref [46] openalexR [47] RISmed [48] ScopusAPI [49] rwos [50] |
| Automated deduplication | ● When new citations are retrieved, a check is performed to see if they already exist in the SOLES database ● Automated deduplication tool identifies duplicates between new citations and existing citations and within new citations (across databases) |
ASySD [39] |
| Machine-assisted screening | ● Trained machine classifier (hosted at EPPI Centre, University College London) determines likelihood of a citation describing experiments in AD animal models | Machine classifiers [38] |
| Automated data annotation | ● Full texts downloaded using open access APIs and publisher (dependent on institutional access) APIs ● Risk of bias reporting assessed using automated tool ● Open access status determined using CrossRef and OpenAlex APIs ● Open data and code status determined by X tool |
rcrossref [46] pre-rob tool [41] openalexR [47] ODDPub [51] |
| Interactive shiny app | ● Shiny application available at https://camarades.shinyapps.io/AD-SOLES/ ● This allows users to: |
|
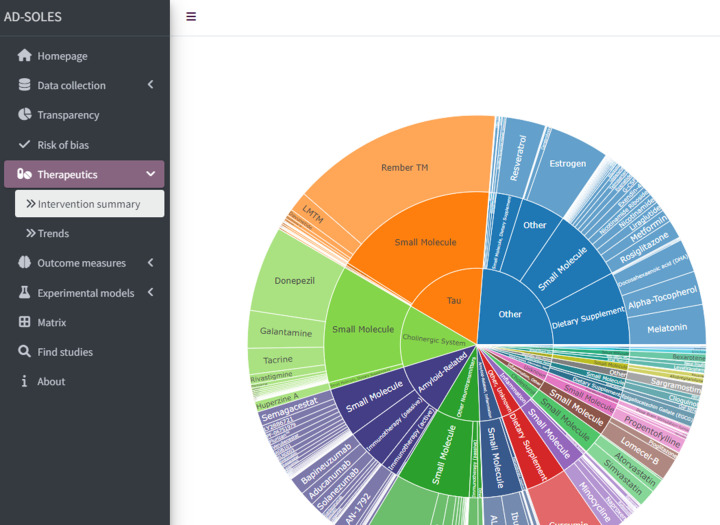
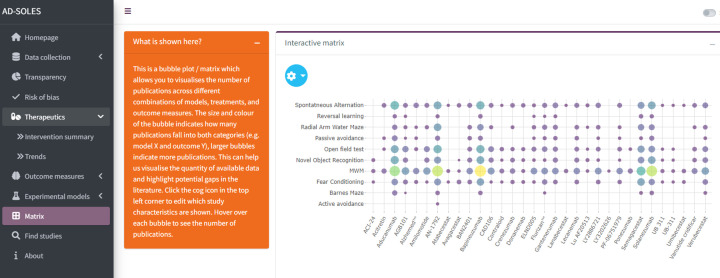
| 1. Gain an overview of the number of relevant publications in different categories, and intersections of those categories | ||
|
||
|
||
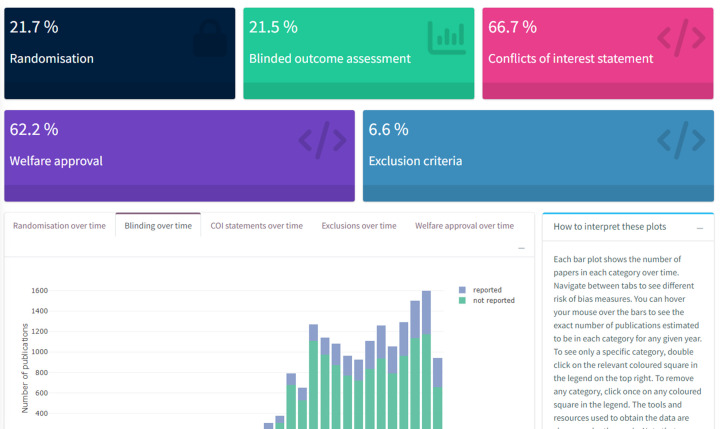
| 2. Visualise the proportion of publications with certain transparency and quality measures over time | ||
|
||
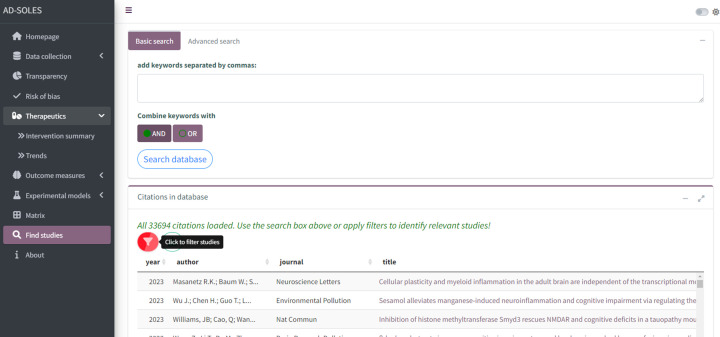
| 3. Download a highly filtered citation lists to start a more detailed systematic literature reviews | ||
|
||
