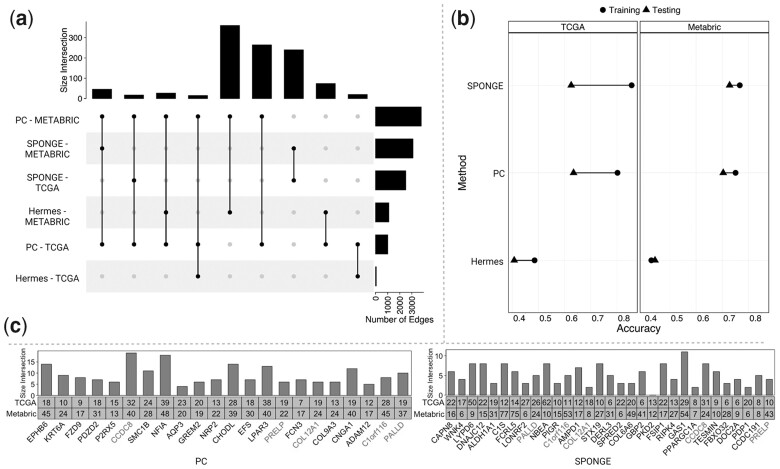
Figure 2.
Testing the robustness of spongEffects modules using the ceRNA network inference methods PC, Hermes, and SPONGE. (a) The overlap of the same edges between the datasets METABRIC and TCGA-BRCA. (b) The accuracy of PC, Hermes, and SPONGE using either METABRIC or TCGA-BRCA as training or independent test set (e.g. if METABRIC is used as a training set, then TCGA-BRCA is used as a test set and vice versa). (c) We compared the top-scoring spongEffects modules (Gini index) from PC and SPONGE. The numbers in the boxes indicate the module size, while the bars indicate the overlap of modules obtained from training on the two different datasets. Red means that the same central ceRNA was found in both methods. We excluded Hermes from this evaluation as it achieved very low prediction performance.