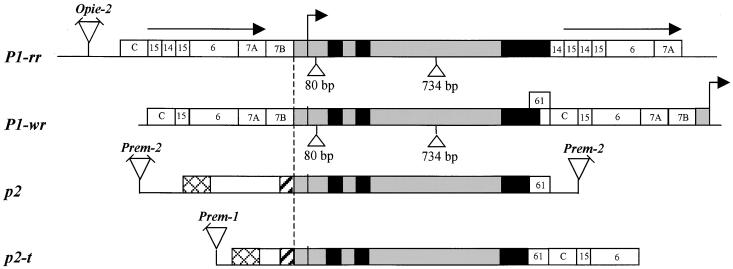
Figure 3.
Structural Comparison among p-Homologous Genes in Maize and Teosinte: Maize P1-rr, Maize P1-wr, Maize p2, and Teosinte p2-t.
The bent arrow indicates the transcription start site of the P1-rr allele (Grotewold et al., 1991a). Black boxes indicate exons, and gray boxes indicate two introns, 5′ UTRs, and 90-bp conserved promoter sequences. Triangles indicate 80- and 734-bp insertions in the P1-rr and P1-wr leader and intron 2, respectively. The P1-rr gene coding region is flanked by 5.2-kb direct repeats indicated by horizontal arrows. Numbered boxes indicate the locations of p1 gene fragments described previously (Lechelt et al., 1989; Athma and Peterson, 1991; Chopra et al., 1996) and used here for homologous sequence comparison. Hatched and cross-hatched boxes indicate homologous sequences (>90%) in the p2 and p2-t genes. Sequences homologous with retroelements are indicated by triangles; angled lines on triangles indicate the absence of an LTR. In the P1-wr allele, a bent arrow at the 3′ end indicates the transcription start of the downstream tandem p1 gene copy. To show structure, the drawing is not to scale. GenBank accession numbers for maize p2 and teosinte p2-t are AF210616 and AF210617, respectively.