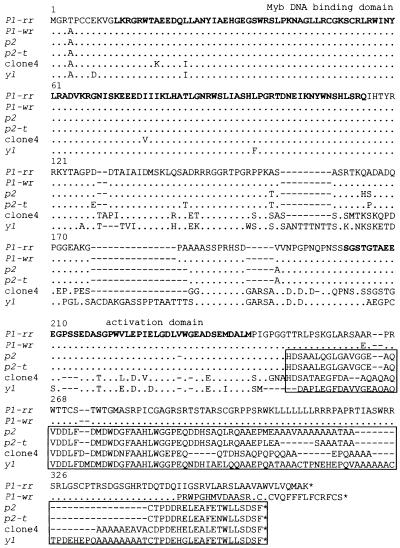
Figure 5.
Alignment of Amino Acid Sequences of Maize, Teosinte, and Sorghum P Proteins.
Amino acid sequences were deduced from the maize P1-rr, maize P1-wr, maize p2, teosinte parviglumis p2-t, teosinte parviglumis clone 4, and sorghum y1 nucleotide sequences. Amino acids of the Myb DNA binding domain and putative transcription activation domain are shown in boldface. Dots indicate identical residues; dashes indicate gaps. The C-terminal boxed regions contain blocks of amino acids conserved among the predicted protein products of the maize p2, teosinte p2-t, teosinte clone 4, and sorghum y1 genes. The P1-rr sequence reported here is derived from the P1-rr-4B2 allele (Grotewold et al., 1991b) and differs at the C-terminal region from the previously reported P1-rr (Bloody Butcher) allele (Grotewold et al., 1991b). The Y1 protein sequence is derived from sorghum genomic (Chopra et al., 1999) and cDNA (S. Chopra and T. Peterson, unpublished data) sequences. The alignments were created by using the Pileup program of Genetics Computer Group software, with subsequent adjustments by inspection to optimize alignments in the C-terminal regions. GenBank accession numbers are as follows: p2, AF210616; p2-t, AF210617; and clone 4, AF210618 and AF210619.