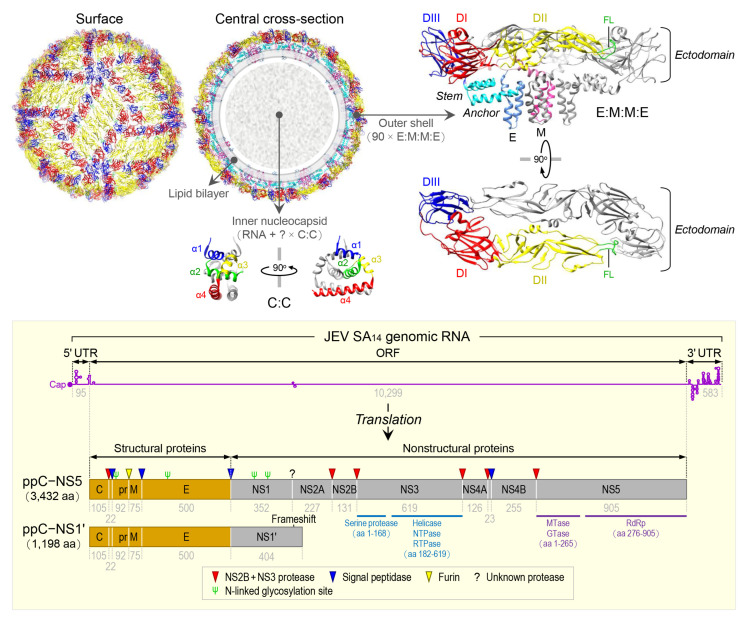
Figure 1.
Virion structure, genome organization, and gene expression of JEV. The top panel shows the high-resolution cryo-electron microscopy structure of JEV strain P3 [102]. The virion contains multiple copies of three proteins: capsid (C), envelope (E), and membrane (M). The C monomer is a cytosolic protein containing four helices (α1, blue; α2, green; α3, yellow; and α4, red). The E monomer is an integral membrane protein comprising three topologically distinct parts: an N-terminal ectodomain, which has three structural domains (DI, red; DII, yellow; and DIII, blue) with the fusion loop (FL, green) positioned at the distal end of DII; a stem (cyan), which has three non-membrane-spanning helices; and a C-terminal anchor (cornflower blue), which has two membrane-spanning helices. The M monomer is also an integral membrane protein comprising three topologically distinct parts: an N-terminal extension containing an unstructured peptide fragment (pink), a non-membrane-spanning helix (hot pink), and a C-terminal anchor containing two membrane-spanning helices (deep pink). The bottom panel depicts the genome organization and gene expression of JEV strain SA14 [103]. The genome is a capped but unpolyadenylated plus-strand RNA, with a single long open reading frame (ORF) flanked by short, highly structured 5’ and 3’ untranslated regions (UTRs). The ORF is translated into two polyprotein precursors, both of which are cleaved by viral and cellular proteases, as indicated, to produce three structural (orange) and seven nonstructural (gray) proteins. Of these proteins, two are multifunctional enzymes: First, NS3 has serine protease, helicase, NTPase, and RTPase activities. Second, NS5 has MTase, GTase, and RdRp activities. Four N-linked glycosylation sites are marked, one in the pr portion of prM, one in E, and two in NS1/1’.