Figure 7.
Functional Complementation and Molecular Characterization of the Arabidopsis chl-6 Mutant.
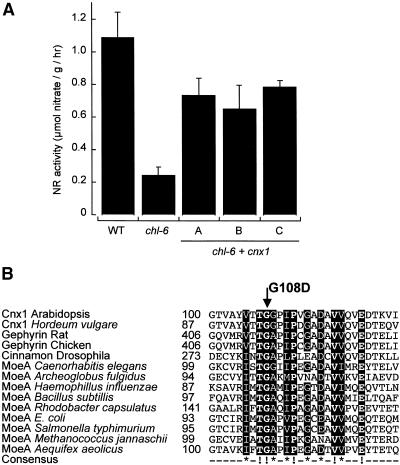
(A) NR activity of the wild-type (WT) and chl-6 mutant plants (B73). Shown are the untransformed mutant (chl-6) and three representative chl-6 plants (A to C) transformed with Arabidopsis cnx1 cDNA (chl-6 + cnx1) under control of the CaMV 35S core promoter (Benfey et al., 1989). Before harvest, plants were grown for 5 days on nitrate-containing media. The bars represent mean values of three to five independent measurements with the standard errors shown.
(B) Alignment of the mutation surrounding the Arabidopsis chl-6 mutation in cnx1. The mutation in the Arabidopsis chl-6 mutant reflects a point mutation at the cDNA position 361 (G → A), resulting in the substitution of aspartate for the glycine 108 (G108D) located in the E domain. Various E domain–homologous sequences were aligned by using ClustalW (www2.ebi.ac.uk/clustalw/); the region surrounding G108D is shown. The numbers shown indicate the first amino acid in each sequence. Protein sequences and the organism from which they are derived are given; GenBank accession numbers are given in the order listed: Q39054, AF268595, Q03555, AF174130, P39205, T20638, A69270, P45210, E69659, AJ23848, P12281, Q56066, Q58296 and E70302. The consensus sequence has been calculated with a threshold of 70%; completely conserved amino acids (boldface letters) are marked (!), and highly conserved residues show an asterisk in the consensus sequence. Both kinds of conserved regions are given in white letters over a black background.