Figure 1.
Genotyping by Array Hybridization.
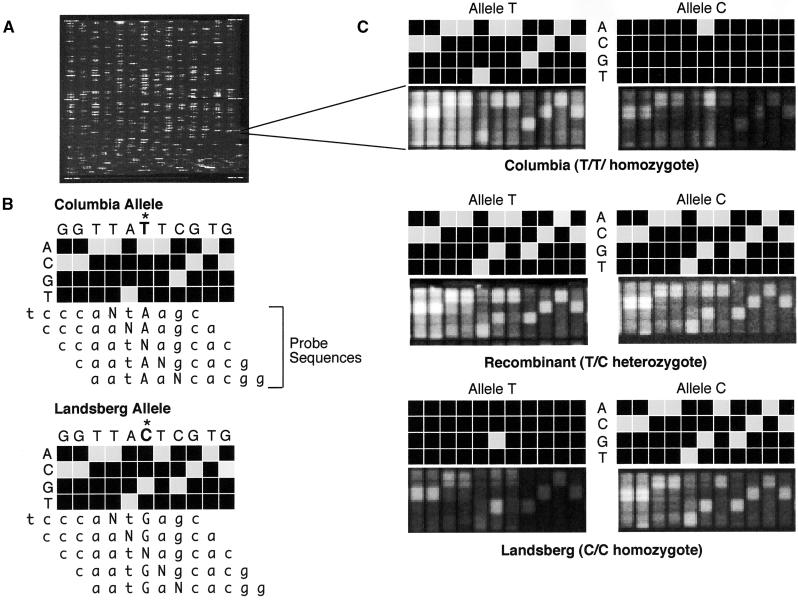
(A) Fluorescence image of an entire oligonucleotide probe array after hybridization.
(B) Scheme of the genotyping of the forward strand of SGCSNP4 in Columbia and Landsberg erecta ecotypes on a variant detector high-density oligonucleotide probe array. VDAs using four 25-mer probes that have an A, C, G, or T at the center position (N) are designed to interrogate not only the polymorphic site (marked with an asterisk) but also the flanking five bases on either side. This design allows determination of the sequence context in which the polymorphic site is embedded and adds to the robustness and accuracy of the genotyping assay. The target DNA hybridizes most strongly to the probe that complements its sequence most closely. Therefore, the probe with the correct base at each center position will produce the strongest hybridization signal.
(C) Scans showing the actual and schematic hybridization patterns for homozygous Columbia (top), homozygous Landsberg erecta (bottom), and a heterozygous recombinant (center). In this example, the variant bases are T and C. Hybridization of the T allele to the C allele VDA, and vice versa, result in only one strong hybridization signal in the column that interrogates the polymorphic site itself. Interrogation of the flanking bases yields either no signals or only weak ones because the target sequence does not match perfectly the corresponding probe sequences, given the different allelic states at the polymorphic site.