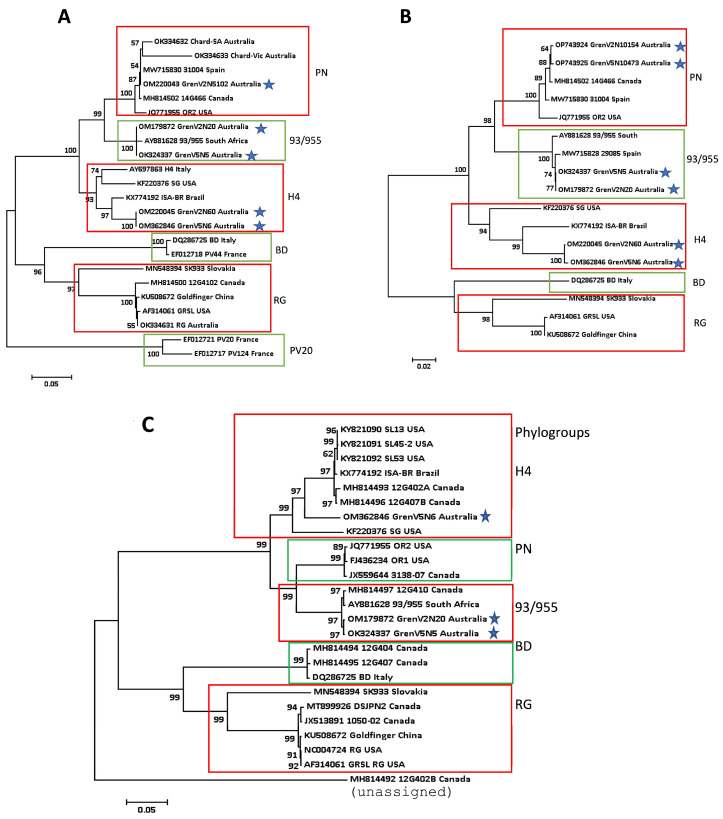
Figure 4.
Phylogenetic mapping of the grapevine leafroll-associated virus 2 (GLRaV-2) variants from Australia and those selected from NCBI. Phylogenetic analysis was carried out using partial nt sequence of CP in Australian-grown Grenache, Chardonnay and Red Globe grapevine varieties. (A) The complete sequence of p24, (B) of selected isolates of GLRaV-2 in Grenache, and (C) nearly full-length virus genome from Vine 2 (V2) and Vine 5 (V5) of Grenache clone SA137. The sequences of the international isolates were obtained from the Genbank NCBI database. The p24 gene sequence of the PV20 phylogroup was not available in the NCBI database. The co-existence of three phylogenetic groups in V2 and V5 is shown by blue asterisks. Neighbour-Joining with bootstrap values of more than 50% (with 1000 repeats) was used for tree construction. Previously classified phylogenetic group names [12] are shown to the right of each clade. For further information on each contig, see Tables S2 and S3. For a clear comparative analysis, the outgroups were not used.