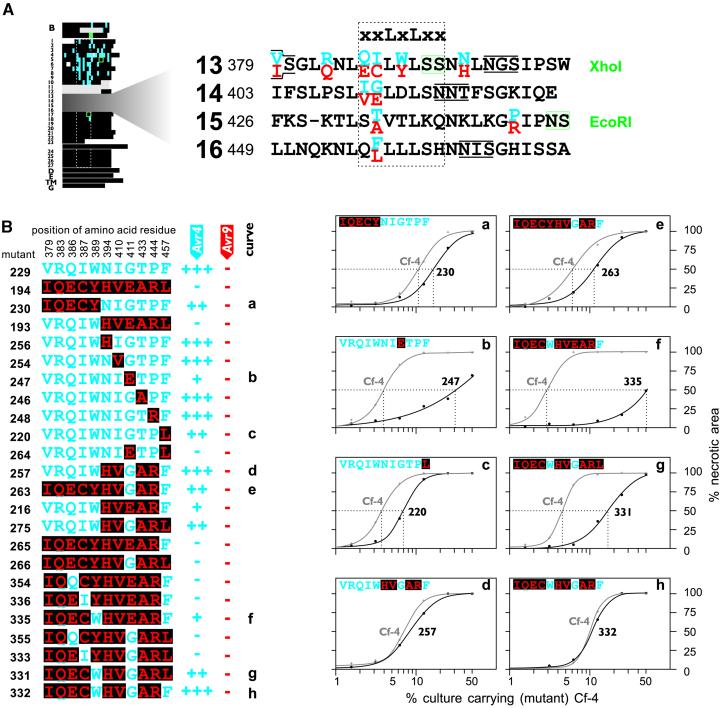
Figure 3.
Three Amino Acid Residues within LRRs 13 to 16 Contribute to Cf-4 Function.
(A) Detailed representation of LRRs 13 to 16. All mutations shown in Figure 3B were introduced into the Cf-4 backbone (left). The enlargement shows an alignment of LRRs 13 to 16 of Cf-4 and Cf-9. Variant amino acid residues between Cf-4 and Cf-9 are shown in blue and red, respectively. Additional details are described in Figure 1.
(B) Identification of single amino acid residues W389, G411, and F457 that contribute to Cf-4 function. Only amino acid residues that differ between Cf-4 and Cf-9 within LRRs 13 to 16 are shown. All mutations were present in the Cf-4 backbone (Figure 3A). Cf-4–specific amino acid residues are shown in blue, and Cf-9–specific amino acid residues are boxed and shown in red. The activity of the various Cf-4 mutants, upon coexpression with either Avr4 or Avr9, is indicated at right: −, no necrosis; +, incomplete necrosis; ++ and +++, complete necrosis. Discrimination between ++ and +++ was determined by quantitative comparisons (curves a to h). For quantitative comparisons, Agrobacterium cultures that carry a plasmid that encodes a (mutant) Cf-4 protein were mixed in different ratios with a culture of equal density carrying an AVR4-encoding plasmid and infiltrated into opposite tobacco leaf halves. At 7 days after infiltration, the percentage of infiltrated area that had become necrotic was measured and plotted against the percentage of culture carrying (mutant) Cf-4.