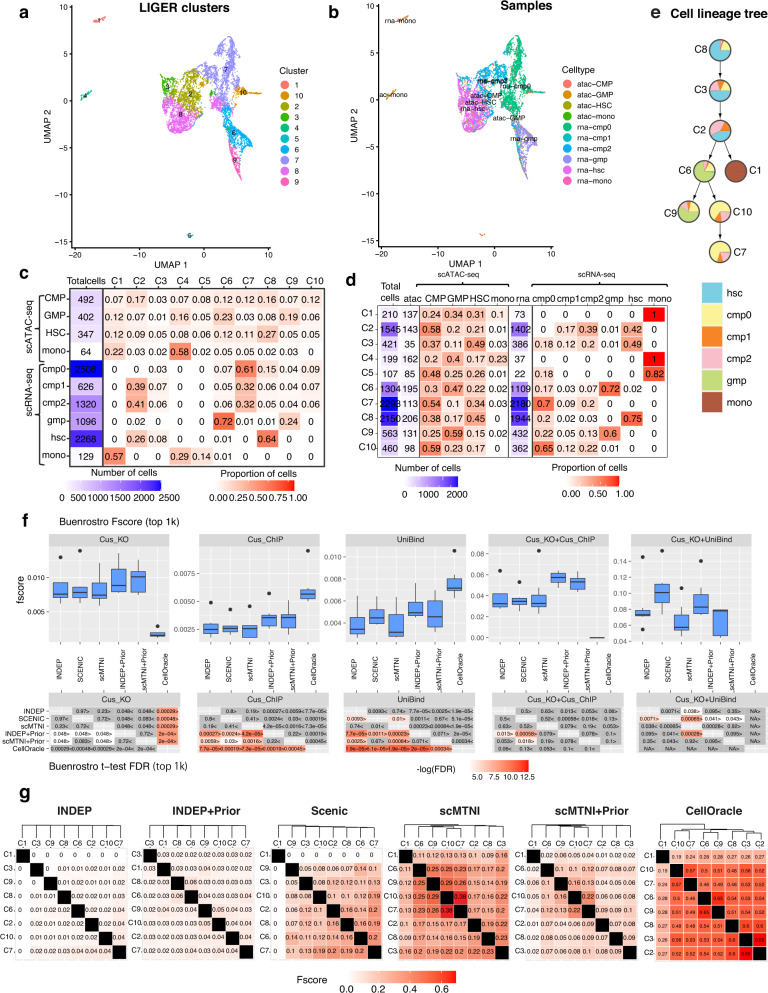
Fig. 5. Inference of cell type-specific networks for human hematopoietic differentiation data.
a UMAP of LIGER cell clusters of the scATAC-seq and scRNA-seq data. b UMAP depicting the original cell types (samples) with scATAC-seq and scRNA-seq data. c The distribution of cell clusters in each sample. d The distribution of samples in each LIGER cluster. e Inferred lineage structure linking the eight cell clusters with scRNA-seq data. f Boxplots showing F-scores of n = 7 cell clusters (all cell clusters excluding C1) for top 1k edges in predicted networks from scMTNI, scMTNI+Prior, INDEP, INDEP+Prior, SCENIC and CellOracle compared to gold standard datasets (top). FDR-corrected t-test to compare the F-score of the row algorithm to the F-score of the column algorithm (bottom). The two-sided paired t-test is conducted on F-scores of n = 7 cell clusters for every pair of algorithms. A FDR < 0.05 was considered significantly better. The sign < or > specifies whether the row algorithm’s F-scores were worse or better than the column algorithm’s F-scores. The color scale is specified by − log(FDR), with the red color proportional to significance. Non-significance is colored in gray. In the boxplot, the horizontal middle line of each plot is the median. The bounds of the box are 0.25 quantile (Q1) and 0.75 quantile (Q3). The upper whisker is the minimum of the maximum value and Q3 + 1.5*IQR, where IQR = Q3 − Q1. The lower whisker is the maximum of the minimum value and Q1 − 1.5*IQR. g Pairwise similarity of networks from each cell cluster using F-score on the top 5k edges. Rows and columns ordered by hierarchical clustering using F-score as the similarity measure. Source data are provided as a Source Data file.