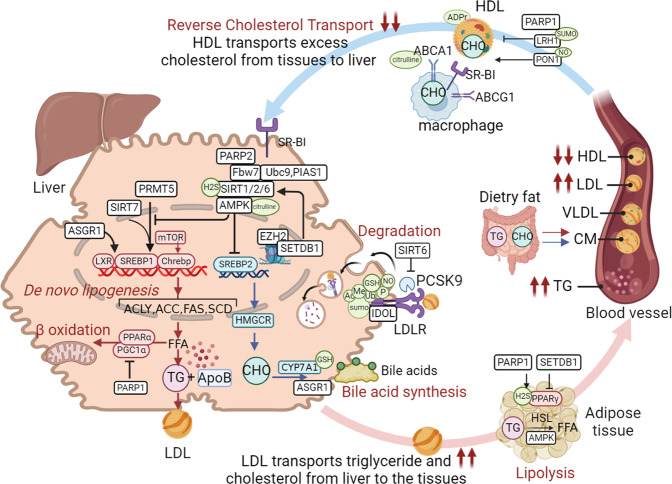
Fig. 6.
Diverse roles of PTMs in hyperlipidemia. Excessive intake and synthesis or insufficient catabolism of lipids lead to hyperlipidemia. De novo lipogenesis (DNL), reverse cholesterol transport (RCT), fatty acid β-oxidation, lipolysis, bile acid synthesis, and lipoprotein-related-regulation contribute to lipid homeostasis at multiple levels. We summarise the role of crosstalk among PTMs in regulating lipid metabolism. (1) During hepatic DNL, abundant PTMs such as phosphorylation (AMPK), (de)acetylation (SIRT1/2/6), methylation (EZH2, PRMT5), ubiquitination (FBW7, ASGR1), SUMOylation (SUMO1, SENP1, UBC9, PIAS1), sulfhydration (NsHS/H2S), citrullination (L-citrulline-AMPK) and ADP ribosylation (PARP2) can regulate the expression of ACLY, ACC, FAS, SCD via transcription factor mTOR, LXR, SREBP and CHREBP. (2) Fatty acid beta-oxidation. ADP ribosylation (PARP1) inhibits beta-oxidation via PPARα. (3) Lipolysis. LDL transports fatty acid from the liver to peripheral tissues. In adipose tissues, acetylation (SETDB1), ADP ribosylation (PARP1), sulfhydration (H2S) and phosphorylation (AMPK) modulate the lipolysis by regulating key enzyme HSL or transcription factor PPARγ. (4) Reverse cholesterol transport (RCT). SUMOylation (SUMO-LRH1), S-nitrosylation (NO-PON1-HDL), citrullination and ADP ribosylation regulate RCT by modifying HDL or relevant enzymes. (5) Bile acid transport. Ubiquitination (ASGR1) and glutathionylation (GSH) govern cholesterol excretion by promoting bile acid synthesis. (6) LDLR. Hepatic LDLR mediates LDL uptake and the subsequent degradation, abolishing excess serum LDL, which is inhibited by PCSK9-mediated LDLR degradation. Several PTMs such as phosphorylation acetylation, methylation, S-nitrosylation, ubiquitination, SUMOylation and glutathionylation could regulate LDLR functioning. Acetylation and ubiquitination modulate LDLR function via the SIRT6-PCSK9-LDLR axis and LXR-IDOL-LDLR axis. The red arrows indicate the triglyceride (TG) pathway, the blue arrows indicate the cholesterol (CHO) pathway, and the black arrows indicate the roles of PTM-related molecules in hyperlipidemia. The figure is generated with BioRender (https://biorender.com). ABCA1 ATP binding cassette transporter, ABCG1 ATP binding cassette transporter, ACC acetyl-CoA carboxylase, ACLY ATP-citrate lyase, AMPK AMP-activated protein kinase, ASGR1 asialoglycoprotein receptor 1, Apo apolipoprotein, CM chylomicrons, CYP7A1 cholesterol 7α-hydroxylase 1, ChREBP carbohydrate-responsive element-binding protein, EZH2 enhancer of zeste homolog 2, FAS fatty acid synthase, FFA fatty acid, GSH glutathione, H2S hydrogen sulfide, HDL high-density lipoprotein, HMGCR 3-Hydroxy-3-methylglutaryl coenzyme A reductase, HSL hormone-sensitive lipase, IDOL inducible degrader of LDLR, LDL low-density lipoprotein, LRH-1 liver receptor homolog-1, LXR liver X receptor, PARP poly (ADP-ribose) polymerase, PGC1α PPARγ coactivator 1α, PIAS1 protein inhibitor of activated STAT 1, PPAR peroxisome proliferator-activated receptor, PRMT protein arginine methyltransferase, SCD stearoyl-CoA desaturase, SENP1 sentrin/SUMO-specific protease 1, SETDB1 SUMOylated SET domain bifurcated 1, SIRT sirtuin, SR-BI scavenger receptor class B member 1, SREBP sterol regulatory element-binding protein, VLDL very low-density lipoprotein, mTOR mammalian target of rapamycin complex, PTMs post translational modifications