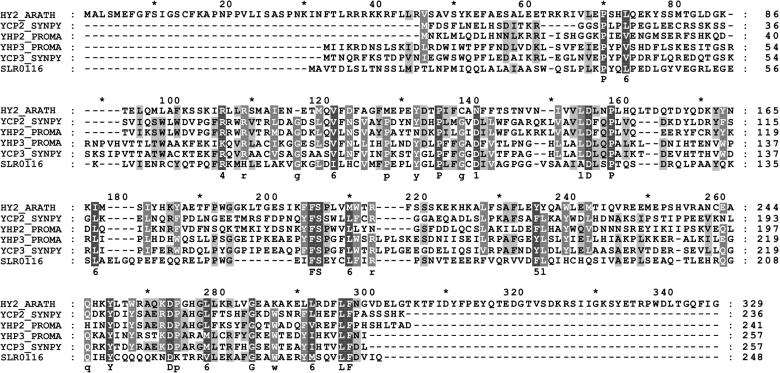
Figure 4.
Alignment of HY2 and HY2-Related Proteins.
Alignment of the HY2 protein with proteins of unknown function from oxygenic photosynthetic bacteria identified by PSI-BLAST. Conserved residues in 100 or 80% of the aligned sequences are depicted in the consensus sequence with uppercase or lowercase letters, respectively. Sequence similarity groups shown in the consensus sequence reflect conservation in >100% of the sequences. These are labeled as follows: 1, 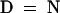 ; 4,
; 4, 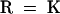 ; 5,
; 5, 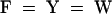 ; and 6,
; and 6, 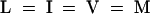 . Dark shading with white letters, gray shading with white letters, and gray shading with black letters reflect 100, 80, and 60% sequence conservation, respectively. Sequence identifiers correspond to hypothetical proteins from Synechococcus sp WH8020 (YCP2_SYNPY and YCP3_SYNPY), from Prochlorococcus (YHP2_PROMA and YHP3_PROMA), and from Synechocystis sp PCC 6803 gene (cyanobase locus slr0116; see http://www.kazusa.or.jp/cyano/cyano.html). Database accession numbers are AB045112 for HY2 (DDBJ), Q02189 for YCP2_SYNPY (SWISS-PROT), Q02190 for YCP3_SYNPY (SWISS-PROT), CAB95700.1 for YHP2_PROMA (EMBL), CAB95701.1 for YHP3_PROMA (EMBL), and S76709 for slr0116 (Protein Information Resource). Asterisks are indicated every 20 residues.
. Dark shading with white letters, gray shading with white letters, and gray shading with black letters reflect 100, 80, and 60% sequence conservation, respectively. Sequence identifiers correspond to hypothetical proteins from Synechococcus sp WH8020 (YCP2_SYNPY and YCP3_SYNPY), from Prochlorococcus (YHP2_PROMA and YHP3_PROMA), and from Synechocystis sp PCC 6803 gene (cyanobase locus slr0116; see http://www.kazusa.or.jp/cyano/cyano.html). Database accession numbers are AB045112 for HY2 (DDBJ), Q02189 for YCP2_SYNPY (SWISS-PROT), Q02190 for YCP3_SYNPY (SWISS-PROT), CAB95700.1 for YHP2_PROMA (EMBL), CAB95701.1 for YHP3_PROMA (EMBL), and S76709 for slr0116 (Protein Information Resource). Asterisks are indicated every 20 residues.