Figure 3: CHK1 functions as a sensor of nuclear H2O2 levels.
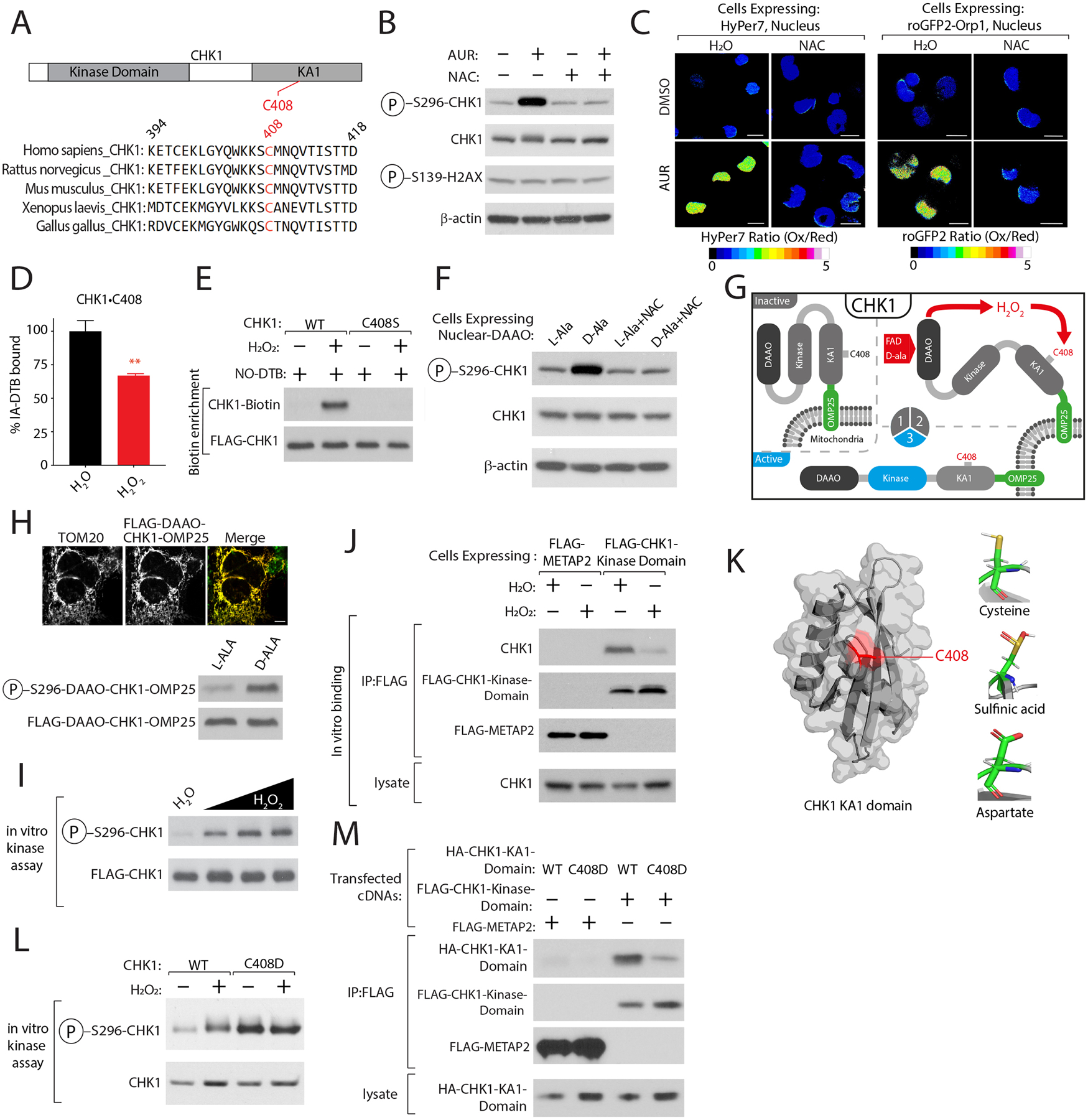
(A) C408 is highly conserved. (B) AUR activates CHK1 in a NAC-dependent manner. K562 cells were treated with AUR in the presence of NAC or vehicle control CHK1 activity was determined by immunoblotting for the indicated proteins. (C) AUR increases the steady-state levels of nuclear H2O2. Ratiometric images of HyPer7 or roGFP2-ORP1 H2O2 reporters localized to the nucleus following indicated treatments (see also Figures S5A–C). (D) H2O2 treatment regulates CHK1•C408 oxidation (see also Table S2B). (E) Oxidation of CHK1•C408 results in sulfinic acid formation (see methods). (F) Nuclear H2O2 activates CHK1. K562 cells stably expressing D-amino acid oxidase (DAAO) localized to the nucleus were treated with either L-Ala or D-Ala or NAC and CHK1 kinase activity was determined as described in (B). (G) Schematic depicting the localization of DAAO-CHK1 to the mitochondrial outer membrane. (H) H2O2 is sufficient to activate CHK1. Top, Immunofluorescence analysis of mitochondrial localization of DAAO-CHK1-OMP25. Bottom, DAAO-CHK1-OMP25 kinase activity was as described in (F). (I) H2O2 directly activates CHK1. CHK1 in vitro kinase assay following treatment with increasing amounts of H2O2. (J) H2O2 reduces the interaction between endogenous CHK1 and the CHK1 kinase domain. (K) Right, Crystal structure of C-terminal Kinase Associate 1 (KA1) domain of CHK1 highlighting the location of C408 in red, adapted from PDB ID: 5WI298. Left, Modeling of CHK1•C408 interactions as a sulfinic acid or when mutated to Asp. (L) CHK1•C408D has elevated kinase activity. (M) CHK1•C408D-mutation in KA1 domain blocks interaction with CHK1 kinase domain. Scale Bar=10 μm. Data are represented as mean ± SEM. **p < 0.001, ***p< 0.0001. Student’s t-test (two-tailed, unpaired) were used to determine statistical significance.
