Figure 5: CHK1 phosphorylates SSBP1 blocking its mitochondrial localization to decrease nuclear H2O2 levels.
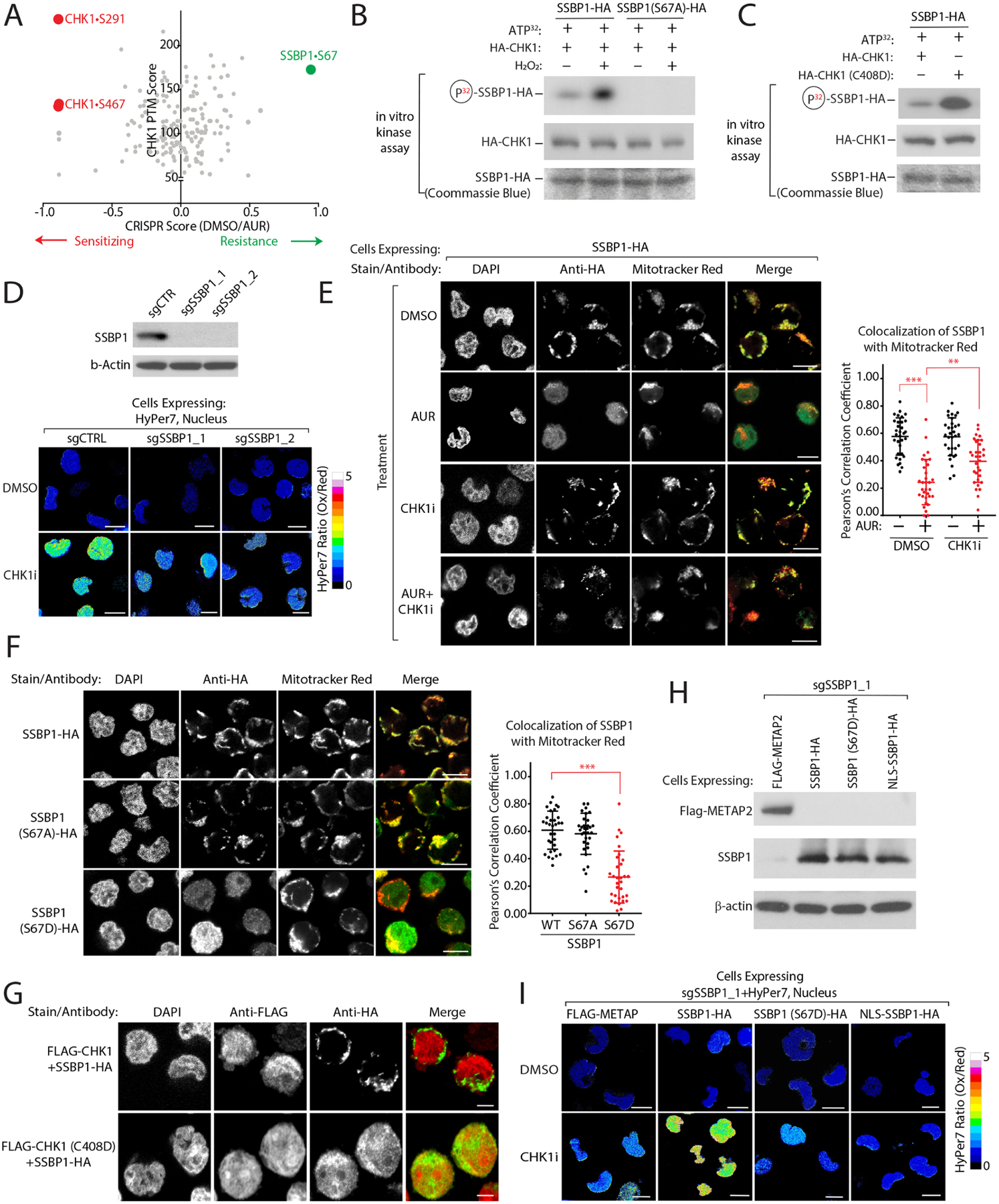
(A) Comparison of CRISPRi scores following AUR treatment with CHK1 phosphorylation sites characterized in Blasius et al.67 identifies SSBP1•S67 as a potential CHK1 target mediating resistance to AUR. (B) SSBP1 is a direct target of CHK1 that is phosphorylated in a H2O2-dependent manner in vitro (see methods). (C) CHK1•C408D has heightened levels of activity towards SSBP1. (D) SSBP1 regulates nuclear H2O2 levels downstream of CHK1. Left, immunoblot of SSBP1 levels in K562-dCas9-KRAB cells expressing the indicated sgRNAs. Right, Measurement of nuclear H2O2 with HyPer7 in K562 cells depleted of the indicated genes. (E) SSBP1 phosphorylation by CHK1 blocks its mitochondrial localization. Left, the localization of SSBP1-HA was determined by immunofluorescence analysis of K562 cells following the indicated treatments. Right, quantification of mitochondrial colocalization of HA-SSBP1. (F) SSBP1•S67D phosphomimetic mutant does not localize to the mitochondria. (G) CHK1•C408D is sufficient to drive SSBP1 re-localization. (H-I) SSBP1•S67D phosphomimetic mutant decreases nuclear H2O2 following CHK1 inhibition. Immunoblot analysis of the indicated proteins reintroduced into SSBP1-depleted cells (H) and corresponding levels of H2O2 levels measured with nuclear HyPer7 (I). Scale Bar=10 μm. Data are represented as mean ± SEM. **p < 0.001, ***p< 0.0001. Statistical significance was determined by Student’s t-test (two-tailed, unpaired).
