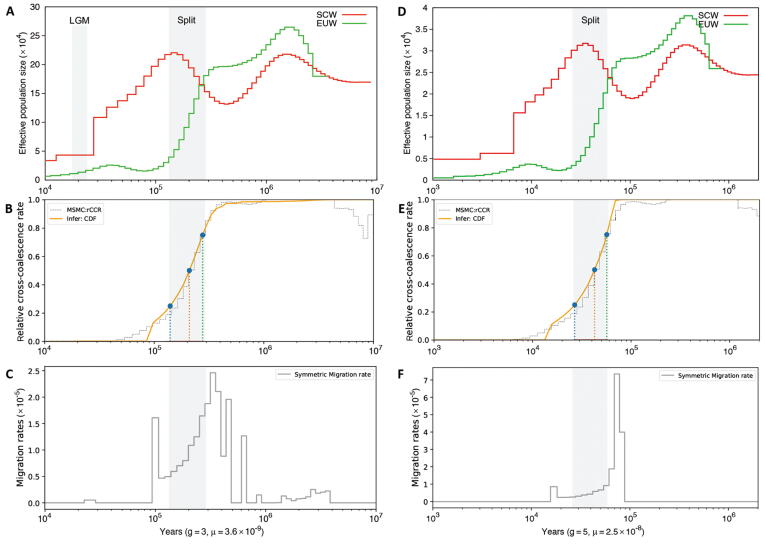
Supplementary Figure S6.
Comparison of population histories inferred using different mutation rates by PSMC, MSMC2, and MSMC-IM The split time is identified and highlighted in grey at the first crossover of population size’s curves instead of the second crossover (backward in time). A and D. Demographic history was inferred with two different mutation rates: 3.6 × 10-9 are the mutation rate estimated in this study; 2.5 × 10-8 are the mutation rate of human, which was widely used in pig demography inference. The LGM is highlighted in grey in (A). B and E. RCCR are inferred by MSMC2 with the two different mutation rates, respectively. The split time was indicated by the RCCR (orange solid line). The vertical dotted lines represented the date corresponding to RCCR of 25%, 50%, and 75%, respectively. C and F. Migration rates are inferred by MSMC-IM with the two different mutation rates, respectively. The split time was reflected via signals of strong gene flow across SCW and EUW. SCW, South Chinese wild pigs; EUW, European wild pigs. LGM, Last Glacial Maximum; PSMC, pairwise sequentially Markovian coalescent model; MSMC2, multiple sequential Markovian coalescent model 2; MSMC-IM, multiple sequential Markovian coalescent-isolation and migration model; RCCR, relative cross-coalescent rates.