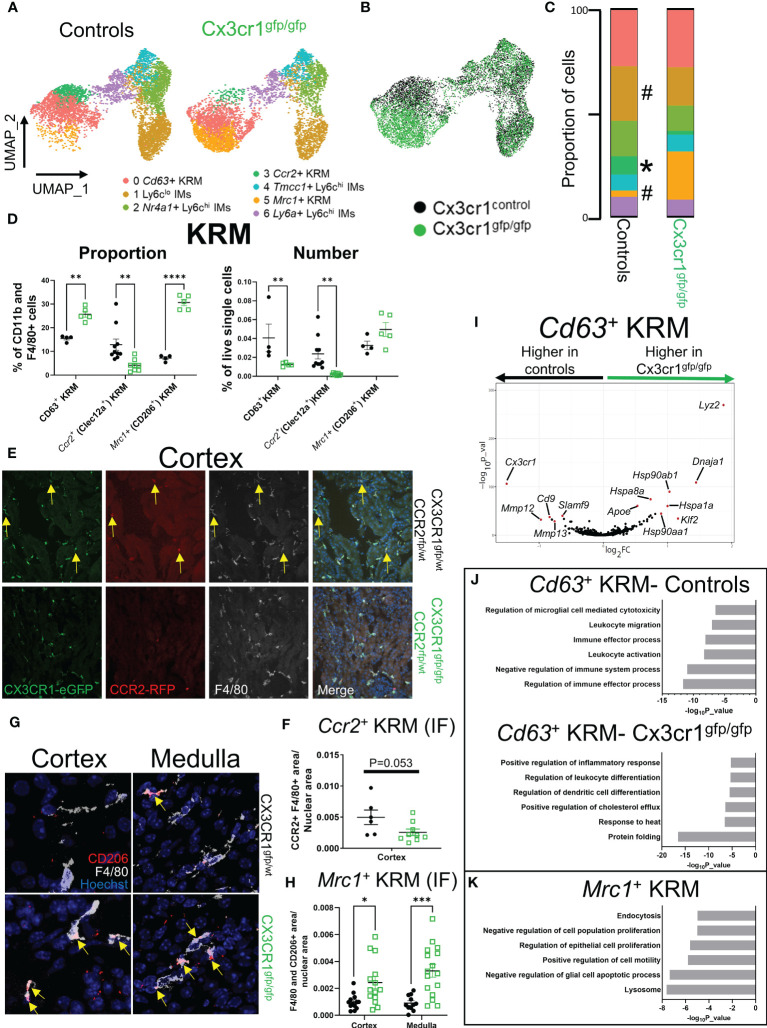
Figure 3.
Cx3cr1 is required for accumulation of Ccr2+ KRM in the kidney cortex. (A) UMAP of cell clusters in control (N=2 mice; both female) and Cx3cr1 gfp/gfp knockout (N=2 mice; both female) mice. The control group shown in this figure is the same as the data shown in Figure 1 . (B) Composite UMAP of cells from control and Cx3cr1 gfp/gfp mice. (C) Quantification of cluster composition. T-test. (D) Quantification of flow cytometry data analyzing KRM subsets as a percentage of CD11b and F4/80 positive cells or as a percentage of live single cells in the kidney. Two-way ANOVA. (E) Representative confocal microscopy images of kidney sections isolated from the cortex of Cx3cr1 gfp/wt Ccr2 rfp/wt (top) or Cx3cr1 gfp/gfp Ccr2 rfp/wt (bottom) stained with the pan macrophage marker F4/80. Mice were 6-8 weeks of age at harvest. N=2-4 mice per group. (F) Quantification of RFP+ F4/80+ area normalized to total nuclear area in Cx3cr1 control or knockout mice. Each dot represents an individual image that was quantified from a combined 2-4 mice per group. T-test. (G) Representative confocal microscopy images of kidney sections isolated from the cortex (left) or medulla (right) of Cx3cr1 gfp/wt (top) or Cx3cr1 gfp/gfp (bottom) mice stained with the pan macrophage marker F4/80 and CD206. Mice were 6-8 weeks of age at the time of harvest. N=3-4 mice per group. (H) Quantification of F4/80+ CD206+ area normalized to total nuclear area in the cortex or medulla of Cx3cr1 control or knockout mice. Each dot represents an individual image that was quantified from a combined 3-4 mice per group. Two-way ANOVA. (I) Volcano plot showing genes that were enriched in Cd63+ KRM isolated from control or Cx3cr1 gfp/gfp kidneys. (J) Metascape pathway analysis of genes that were significantly enriched (adjusted p value < 0.05) in Cd63+ KRM isolated from control or Cx3cr1 gfp/gfp kidneys. (K) Metascape pathway analysis of genes that were significantly enriched (adjusted p value < 0.05) in Mrc1+ KRM compared to Cd63+ KRM. #P <0.1, *P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001.