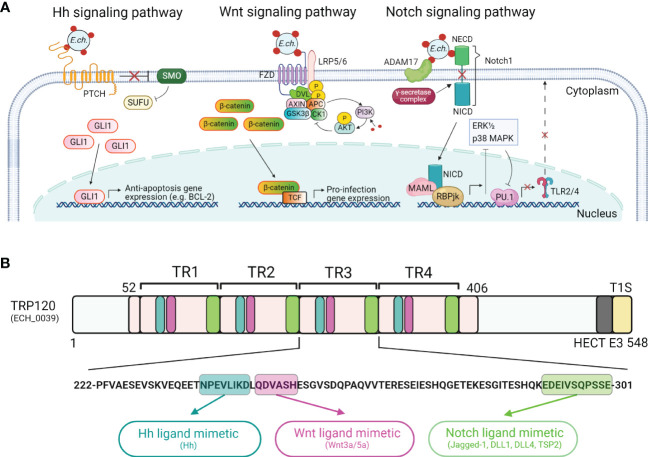
Figure 2.
Ehrlichia SLiM ligand mimicry activation of conserved signaling pathways. (A) Proposed model of E. chaffeensis manipulation of Hedgehog (Hh), Wnt, and Notch signaling pathways by T1SS effector, TRP120. For the Hh signaling pathway, dense-core E. chaffeensis surface-expressed TRP120 binds to the Hh receptor PTCH to activate the GLI1 zinc finger transcription factor. GLI1 translocation to the nucleus activates the transcription of the anti-apoptosis target genes such as BCL-2. In the canonical Wnt signaling pathway, E. chaffeensis TRP120 directly engages FZD (Frizzled) 5 and recruits coreceptor LRP5/6 (lipoprotein receptor-related protein 5 and 6). Activation of Wnt signaling results in the disassembly of the β-catenin destruction complex [consisting of Axin, APC (adenomatous polyposis coli), GSK3β (glycogen synthase kinase 3 beta), and CK1 (casein kinase)], which allows accumulation of β-catenin in the cytoplasm and subsequent nuclear translocation and activation of Wnt target pro-infection genes and Wnt signaling promotes PI3K (phosphatidylinositol 3-kinase)/AKT signaling and downstream suppression of autophagy. In the Notch signaling pathway, E. chaffeensis TRP120 interacts with ADAM17 (a disintegrin and metalloproteinase 17) and the Notch1 receptor, resulting in receptor cleavage and nuclear translocation of NICD (Notch intracellular domain), the transcriptionally active form that interacts with Notch RBPjk (recombinant binding protein suppressor of hairless involved in Notch signaling) and MAML (mastermind-like protein 1) transcription factors proteins. This transcription complex then activates transcription of Notch target genes, resulting in inhibition of ERK1/2 (extracellular signal-regulated kinases) and p38 MAPK (p38 mitogen-activated protein kinase) phosphorylation pathway and the downstream transcription factor PU.1 expression is repressed, inhibiting TLR2/4 expression. (B) E. chaffeensis TRP120 activates conserved signaling pathways via TRP120 Hh-, Wnt- and Notch-specific SLiM ligand mimetics embedded within each TR domain. Combined bioinformatic and functional characterization have identified E. chaffeensis TRP120 Hh, Wnt and Notch repetitive SLiMs as shown in blue (Hh), purple (Wnt), and green (Notch), respectively.