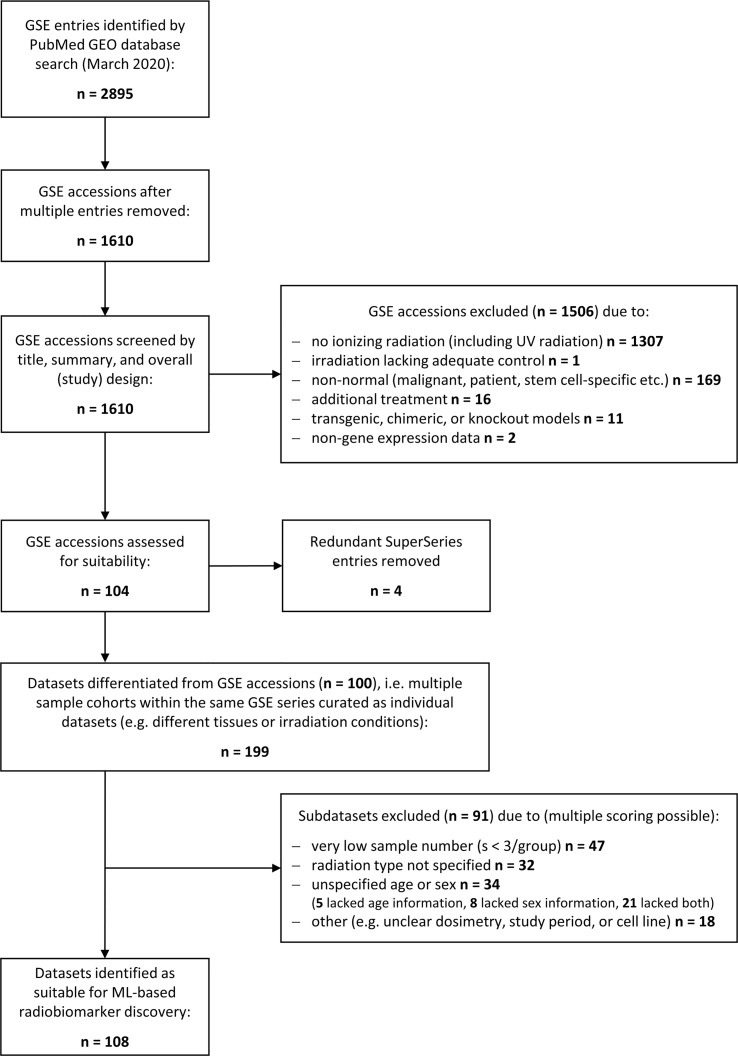
Figure 1.
Schematic illustrating the database search to identify gene expression datasets for normal tissue radiation responses. Flowchart showing the database search performed at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) resource (https://www.ncbi.nlm.nih.gov/geo/). The search was performed in March 2020 yielding 2895 transcriptional gene expression entries (mRNA, miRNA, lncRNA etc.) that consisted to a large part of multiple entries. The remaining 1610 unique accessions were screened to identify normal tissue datasets from any mammalian species subjected to any type of ionizing radiation (X-rays, alpha or beta particles, heavy ions, etc.), any source (external beams, (partially) sealed radioactive sources, or injected radionuclides), and any form of exposure (external, internal, whole body or partial body irradiation, etc.). Most of the accession numbers were unsuitable for normal tissue biomarker identification and excluded. The remaining accessions were manually curated to delineate subsets of data with insufficient data quality such as low sample number, unspecified dosimetry, or time points. In total, 108 suitable datasets for ML processing were identified.