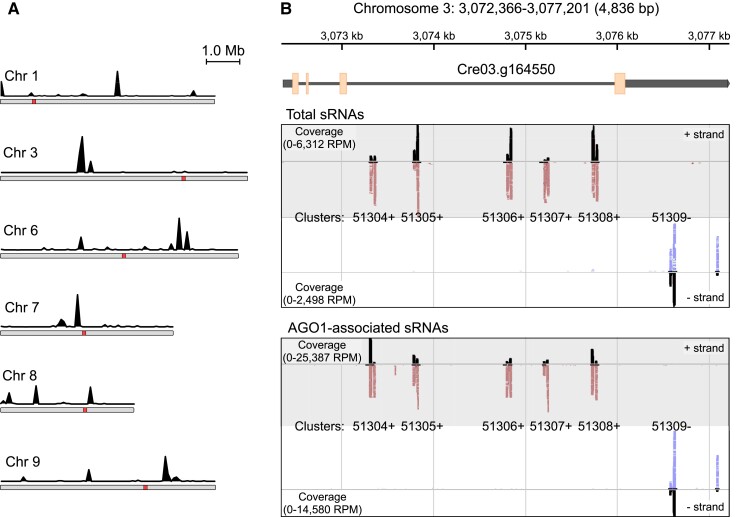
Figure 4.
AGO1-associated >26-nt sRNAs originate from clustered genomic loci. A) Location of genomic loci producing AGO1-associated >26-nt sRNAs on Chlamydomonas chromosomes. The black traces indicate the number of AGO1-associated long sRNAs (from normalized libraries) mapped in sliding 10-kb windows across the Chlamydomonas chromosomes. Red squares represent putative centromeres (Craig et al. 2021). B) Genome browser view of the clusters matching to the Cre03.g164550 gene, encoding a homolog of the conserved eukaryotic FRA2/BolA-like protein 2. The upper panel shows the chromosome 3 coordinates and a diagram of the Cre03.g164550 gene with exons (orange boxes), introns (thin gray lines), and UTRs (thick gray lines). The middle panel shows sRNAs from total sRNA libraries mapping to the sense strand (upper) or the antisense strand (lower). Individual mapped sRNAs are shown as red marks (mapped on + strand) or blue marks (mapped on − strand) along with coverage in black (indicated as RPM). The bottom panel shows AGO1-associated >26-nt sRNAs mapping to the sense strand (upper) or the antisense strand (lower).