Abstract
The CLASS III HOMEODOMAIN-LEUCINE ZIPPER (HD-ZIPIII) transcription factors (TFs) were repeatedly deployed over 725 million years of evolution to regulate central developmental innovations. The START domain of this pivotal class of developmental regulators was recognized over 20 years ago, but its putative ligands and functional contributions remain unknown. Here, we demonstrate that the START domain promotes HD-ZIPIII TF homodimerization and increases transcriptional potency. Effects on transcriptional output can be ported onto heterologous TFs, consistent with principles of evolution via domain capture. We also show the START domain binds several species of phospholipids, and that mutations in conserved residues perturbing ligand binding and/or its downstream conformational readout abolish HD-ZIPIII DNA-binding competence. Our data present a model in which the START domain potentiates transcriptional activity and uses ligand-induced conformational change to render HD-ZIPIII dimers competent to bind DNA. These findings resolve a long-standing mystery in plant development and highlight the flexible and diverse regulatory potential coded within this widely distributed evolutionary module.
The START domain potentiates HD-ZIPIII TF activity and uses ligand-induced conformational change to enable DNA binding, a finding that answers a long-standing question in plant development.
IN A NUTSHELL.
Background: Development has been compared to a ball rolling down a hill. Cells initially have broad potential, but over time, they make decisions based on local cues which set them down a path to differentiation. Along this trajectory, thousands of genes need to be turned off or on in a carefully choreographed manner to ensure specialized identity. This is accomplished in part by transcription factors whose activities are precisely controlled by inputs operating across multiple regulatory levels. Here, we study the regulation of CLASS III HOMEODOMAIN LEUCINE ZIPPER (HD-ZIPIII) proteins, a 725-million-year-old family of transcription factors that were redeployed throughout evolution to impact nearly all aspects of plant development.
Question: HD-ZIPIII proteins contain a START domain, an evolutionarily ubiquitous module that binds lipophilic ligands. The hypothesis thus arose that HD-ZIPIII activity may be controlled by lipid ligand inputs. Remarkably, both the ligands and regulatory properties of the HD-ZIPIII START domain have remained unknown for over 20 years. The goal of our study was to resolve this long-standing mystery.
Findings: We find that the START domain promotes HD-ZIPIII dimerization and increases their transcriptional potency. We also identify several phospholipid ligands bound by the HD-ZIPIII START domain, and show that perturbing ligand binding abolishes DNA-binding competence. Thus, the START domain turns HD-ZIPIII proteins into potent DNA-binding competent transcription factors but only if they can bind and respond to their phospholipid ligands. Our findings further highlight the remarkably flexible and diverse regulatory potential of START domains.
Next steps: Having established START regulatory properties for an HD-ZIPIII protein in Arabidopsis, we are now considering our findings from an evolutionary perspective. How and when did these regulatory properties come about? Is there functional divergence of START regulation of HD-ZIPIII activity? And how are START domains able to employ such a diverse set of regulatory mechanisms?
Introduction
Development of multicellular organisms requires the precise control of transcription factor (TF) inputs into their gene regulatory networks. As such, the activity of TFs is highly regulated, often integrating distinct mechanisms across multiple regulatory levels to impact developmental outcomes. In plants, this is exemplified by CLASS III HOMEODOMAIN-LEUCINE ZIPPER (HD-ZIPIII) proteins, an ancient TF family that arose after the divergence of unicellular Chlorophyta but before the emergence of Streptophyte algae and land plants over 725 million years ago (Ariel et al. 2007; Goodstein et al. 2012; Romani et al. 2018). HD-ZIPIII TFs have been repeatedly co-opted throughout plant evolution to regulate key developmental advances (McConnell et al. 2001; Juarez et al. 2004; Prigge et al. 2005; Kelley et al. 2009; Carlsbecker et al. 2010; Robischon et al. 2011; Sebastian et al. 2015; Yip et al. 2016; Ramachandran et al. 2017; Xu et al. 2019). For instance, in Arabidopsis thaliana, HD-ZIPIII genes contribute to vascular specification (Carlsbecker et al. 2010; Ramachandran et al. 2017; Smetana et al. 2019), root and shoot apical meristem maintenance (Prigge et al. 2005; Sebastian et al. 2015; Ramachandran et al. 2017), and the distinction of adaxial tissues in lateral organs (McConnell et al. 2001; Prigge et al. 2005). These innovations parallel the increasing complexity of plant form and were instrumental to the enormous success of land plants (Ariel et al. 2007; Goodstein et al. 2012; Romani et al. 2018).
In keeping with a critical role in development, HD-ZIPIII activity is subject to intricate regulation. HD-ZIPIII transcripts are targeted by small RNAs of the miR166 family, which restricts their accumulation via morphogen-like patterning properties (Skopelitis et al. 2017). Loss of this regulation conditions gain-of-function phenotypes impacting nearly all aspects of plant development (McConnell et al. 2001). At the protein level, HD-ZIPIII activity is modulated in part through interaction with their direct targets, the LITTLE ZIPPER (ZPR) family. ZPR proteins capture HD-ZIPIII TFs into heteromeric complexes lacking DNA-binding potential, creating a negative feedback loop that fine-tunes HD-ZIPIII activity (Wenkel et al. 2007; Kim et al. 2008; Husbands et al. 2016).
An additional layer of regulation is suggested by the fact that HD-ZIPIII proteins contain a START domain (Fig. 1A, Ponting and Aravind 1999). START domains are members of the StARkin superfamily (derived from “kin of steroidogenic acute regulatory protein (StAR)”), which are present throughout the tree of life (Wong and Levine 2016; Dresden et al. 2021). StARkin domains are characterized by an α/β helix-grip fold structure with a deep hydrophobic pocket that accommodates lipophilic ligands such as long-chain fatty acids, sterols, and isoprenoids (Tsujishita and Hurley 2000; Roderick et al. 2002; Hubbard et al. 2010). Ligand binding induces stereotypical conformational changes that activate StARkin proteins through a diverse set of non-mutually exclusive regulatory mechanisms (reviewed in Dresden et al. 2021). For instance, StARkin domains can control protein turnover, homomeric and heteromeric complex stoichiometry, subcellular localization, and secondary structure stability (Alpy and Tomasetto 2005; Kanno et al. 2007; Park et al. 2009; Du et al. 2012; Schrick et al. 2014; Belda-Palazon et al. 2018; Zhang et al. 2018; Iida et al. 2019; Tillman et al. 2020; Nagata et al. 2021). The presence of a homeodomain and a START domain therefore sparked a long-standing hypothesis that the transcriptional activity of HD-ZIPIII proteins may be controlled by a lipid ligand, in a manner reminiscent of nuclear receptors in mammalian systems (Schrick et al. 2004; Lumba et al. 2010; Sladek 2011). Remarkably, the role of the HD-ZIPIII START domain remains unclear, despite the essential roles these TFs play in development (McConnell et al. 2001; Prigge et al. 2005).
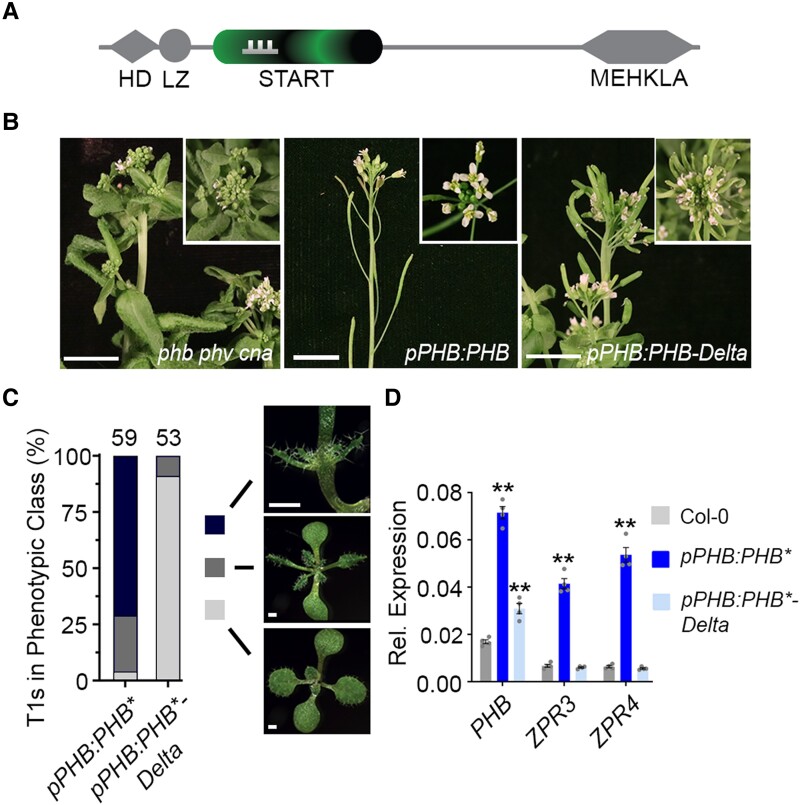
Figure 1.
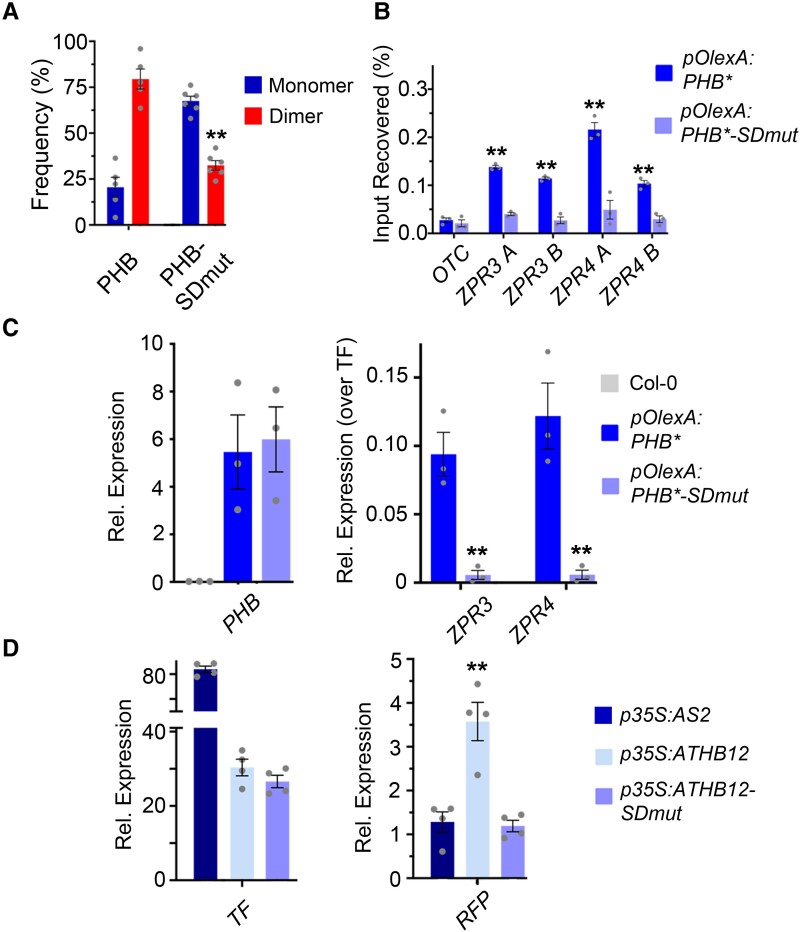
The START domain is required for full PHB function. A) General structure of HD-ZIPIII proteins. Note PHB-Delta retains the miR166-binding site (comb) contained within the START domain. B) The pleiotropic phb phv cna triple mutant phenotype has been previously described (Prigge et al. 2005) and includes fasciated stems and abnormal floral organs (left). The pPHB:PHB transgene complements the mutant phenotype and plants appear wild-type (middle), whereas pPHB:PHB-Delta plants do not (right). C) Phenotypic scoring of primary transformants (n above bar) carrying miR166-insensitive (*) pPHB:PHB or pPHB:PHB-Delta constructs. Ectopic accumulation of PHB leads to severe or intermediate gain-of-function phenotypes, whereas plants mis-accumulating PHB-Delta appear wild-type. D) Ectopic PHB* expression leads to strong upregulation of ZPR3 and ZPR4 targets. By contrast, ZPR3 or ZPR4 levels are indistinguishable from Col-0 in pPHB:PHB*-Delta lines, despite accumulation of this variant above endogenous PHB levels. Note: higher PHB transcript levels in pPHB:PHB* could reflect either a PHB auto-activation mechanism (proposed in McConnell et al. 2001) or the greater relative proportion of adaxialized tissues in pPHB:PHB* seedlings (versus pPHB:PHB*-Delta). n = 3 biological replicates. **P ≤ 0.01, Student's t-test.
Potential insights may come from the evolutionarily related HD-ZIPIV family, which also diverged at least 725 million years ago (Ariel et al. 2007; Goodstein et al. 2012; Romani et al. 2018), and whose START domain is required for function (Schrick et al. 2014; Iida et al. 2019; Nagata et al. 2021). Yeast expression analyses indicate that this START domain binds a broad spectrum of metabolites and increases TF stability (Schrick et al. 2014). Recent studies find similar promotive effects on protein stability in plants and identify subcellular localization as an additional HD-ZIPIV START regulatory mechanism controlling epidermal cell fate (Iida et al. 2019; Nagata et al. 2021). These effects are thought to be mediated by binding of epidermally synthesized ceramides, which would reinforce their tissue-specific activity (Iida et al. 2019; Nagata et al. 2021). Given the long evolutionary divergence of HD-ZIPIII proteins (Ariel et al. 2007; Goodstein et al. 2012; Romani et al. 2018), their functions outside of the epidermis (McConnell et al. 2001; Juarez et al. 2004; Prigge et al. 2005; Kelley et al. 2009; Carlsbecker et al. 2010; Robischon et al. 2011; Sebastian et al. 2015; Yip et al. 2016; Ramachandran et al. 2017; Xu et al. 2019), and the fact that HD-ZIPIII and HD-ZIPIV START domains are not interchangeable in yeast or plant assays (Schrick et al. 2014), the extent to which these observations apply to the HD-ZIPIII START domain is difficult to predict.
We show that addition of the HD-ZIPIII START domain potentiates transcriptional activity by promoting homodimerization and increasing transcriptional potency. Further, mutations that affect ligand binding and likely its downstream conformational response abolish DNA-binding competence, without overt effects on protein stability, subcellular localization, and interaction partners. Thus, the HD-ZIPIII START domain potentiates TF activity through regulatory mechanisms distinct from the evolutionarily related HD-ZIPIV START domain. These findings resolve a long-standing mystery in plant development and highlight the flexible and diverse regulatory potential coded within the ubiquitously distributed StARkin evolutionary module.
Results
The START domain is required for full PHB function
We assessed START-dependent effects on HD-ZIPIII developmental function using Arabidopsis thaliana PHABULOSA (PHB) as a representative member. We began by replacing the PHB START domain in a functional YFP-tagged pPHB:PHB reporter (Skopelitis et al. 2017) with the 21-nt microRNA166 (miR166) recognition site found within the START domain coding sequence (pPHB:PHB-Delta, Rhoades et al. 2002). Whereas the pPHB:PHB transgene complements the phb, phavoluta (phv), and corona (cna) triple mutant phenotype, the pPHB:PHB-Delta construct fails to rescue (Fig. 1B), despite equivalent accumulation of PHB and PHB-Delta transcripts (Supplemental Fig. S1A). We therefore next introduced a silent mutation into the 21-nt miR166 binding site, abolishing miR166 regulation of PHB and PHB-Delta (pPHB:PHB*; pPHB:PHB*-Delta). Loss of miR166 regulation generates a highly sensitive, dosage-dependent readout of HD-ZIPIII activity (McConnell et al. 2001; Mallory et al. 2004; Carlsbecker et al. 2010; Skopelitis et al. 2017), permitting detection of weak or subtle HD-ZIPIII function that might be missed in standard complementation assays. As expected (Mallory et al. 2004), over 90% of miR166-insensitive pPHB:PHB* primary transformants show PHB gain-of-function phenotypes (Fig. 1C). By contrast, pPHB:PHB*-Delta transformants are indistinguishable from wild-type plants (Fig. 1C), despite similar ectopic accumulation of protein (Supplemental Fig. S1B to E). Further, transcript levels of the HD-ZIPIII direct targets ZPR3 and ZPR4 are strongly upregulated in pPHB:PHB* lines but are indistinguishable from wild-type in pPHB:PHB*-Delta transformants (Fig. 1D). Thus, the START domain is required for PHB to fulfill its developmental function. Moreover, as PHB-Delta shows no obvious effects on protein stability or subcellular localization (Supplemental Fig. S1B to E, and see below), the HD-ZIPIII START domain employs regulatory mechanisms distinct from those of the evolutionary related HD-ZIPIV START domain.
The START domain promotes PHB dimerization
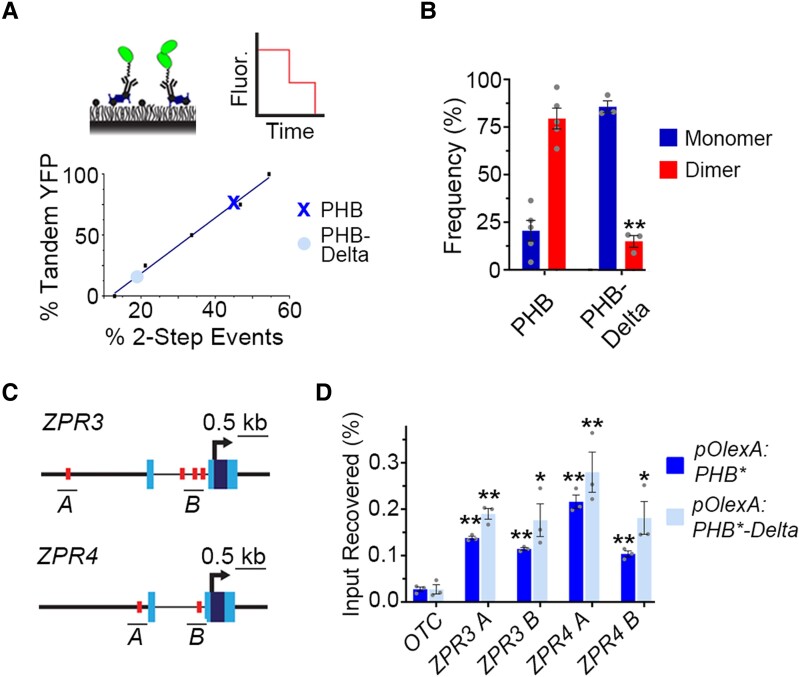
One frequently used StARkin regulatory mechanism is modulation of homomeric stoichiometry (Fujii et al. 2009; Yin et al. 2009; Hubbard et al. 2010; Gatta et al. 2015; Prashek et al. 2017; Tillman et al. 2020; Sanchez-Solana et al. 2021). To test whether the START domain impacts PHB homodimerization, we used single-molecule pull down (SiMPull), which we previously adapted for plant systems (Husbands et al. 2016). We first determined the maturation frequency of the monomeric citrine yellow fluorescent protein (YFP) variant in Arabidopsis to calibrate the frequency of two-step photobleaching events into a quantitative assessment of homomeric stoichiometry (Fig. 2A and Supplemental Fig. S2, Jain et al. 2011, 2014; Aggarwal and Ha 2014; Husbands et al. 2016). Subsequent analyses of over 3,300 protein complexes showed that ∼80% of wild-type PHB proteins are present as dimers (Fig. 2B). This frequency resembles that of strongly homodimeric proteins in animal systems (Jain et al. 2011, 2014), and predicts PHB functions primarily as a homodimer. By contrast, PHB-Delta has a dimerization frequency of 15%—about twice the frequency with which two-step photobleaching events are observed by chance (Fig. 2B and Supplemental Fig. S2B). Thus, the PHB START domain promotes either the formation or the maintenance of PHB homodimers (Fig. 2B). As HD-ZIPIII proteins require dimerization to bind DNA (Sessa et al. 1998), this effect on homodimerization provides a potential explanation for PHB-Delta failing to activate the normal PHB developmental program (Fig. 1B to D).
Figure 2.
The START domain affects PHB homodimerization. A) SiMPull calibration curve (bottom graph) for monomeric vs dimeric YFP (top left cartoon) translates percentage of two-step photobleaching events (simulated in top right cartoon) to frequency of dimers in the population. PHB (X) and PHB-Delta (●) show two-step photobleaching events of 47% and 20%, respectively. B) Percentages of two-step photobleaching events translate to dimerization frequencies of ∼80% for PHB and ∼15% for PHB-Delta. C) Schematic representations of ZPR3 and ZPR4 showing HD-ZIPIII binding sites identified by FIMO (red boxes), ChIP amplicons (black bars), transcription start sites (arrow), untranslated regions (light-blue boxes), and exons (dark-blue boxes). D) PHB and PHB-Delta occupy multiple sites in the regulatory regions of ZPR3 and ZPR4 and are significantly enriched over the ORNITHINE TRANSCARBAMYLASE (OTC) negative control locus. n = 3 biological replicates. *P ≤ 0.05, **P ≤ 0.01, Student's t-test.
The START domain enhances PHB transcriptional potency
One consequence of poor dimerization of PHB-Delta is that it may obscure possible additional contributions from the START domain to HD-ZIPIII TF function. We therefore turned to a short-term estradiol-inducible overexpression system to increase the total number of PHB-Delta dimers available for molecular assays of TF activity. Importantly, estradiol induction does not change the subcellular localization or dimerization frequencies of PHB or PHB-Delta seen at native expression levels (Supplemental Fig. S3A to D). Both variants can also be induced to similar levels and show comparable protein stability (Supplemental Fig. S3, A and E), making this approach suitable for identifying additional START-dependent effects.
Using short-term estradiol-inductions, we first tested whether PHB proteins lacking the START domain are capable of binding to DNA using chromatin immunoprecipitation (ChIP). PHB occupies multiple palindromic HD-ZIPIII binding sites (Sessa et al. 1998) in the regulatory regions of its ZPR3 and ZPR4 direct targets (Fig. 2, C and D). Similarly, enrichment at these sites was also detected for PHB-Delta (Fig. 2, C and D), indicating this variant retains the capacity to bind to DNA.
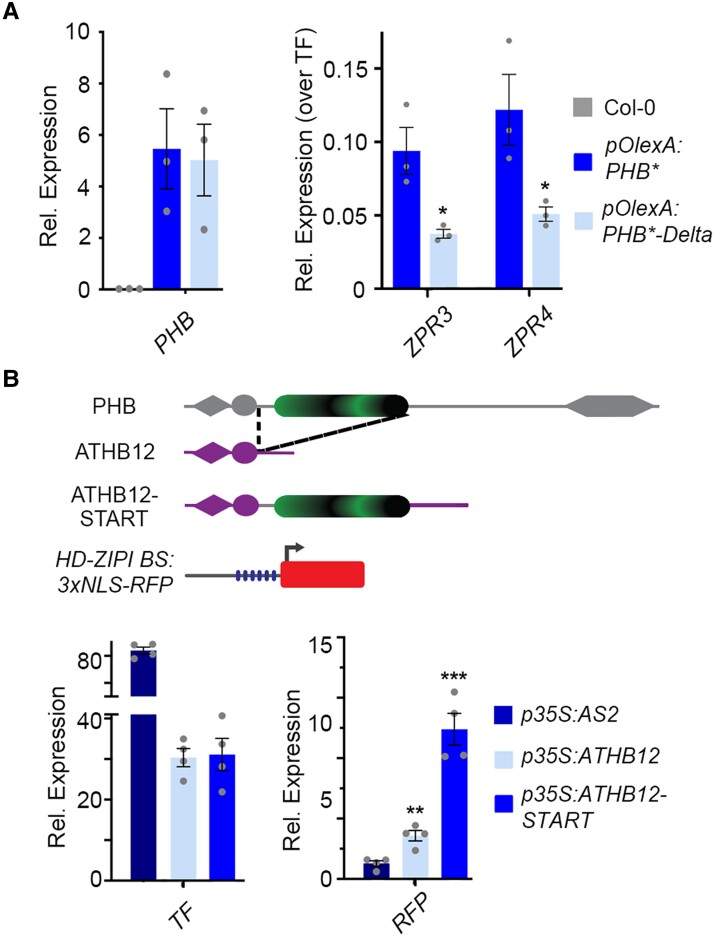
Given this outcome, we next gauged the effect of the START domain on transcriptional potency. Here, the short-term estradiol-induction system has the added benefit that it provides a direct quantitative readout of transcriptional activity while avoiding confounding consequences of morphological changes and regulatory feedback (Wenkel et al. 2007; Kim et al. 2008). As expected, ZPR3 and ZPR4 transcript levels are strongly upregulated upon induction of PHB (Fig. 3A). ZPR3 and ZPR4 targets are also upregulated upon induction of PHB-Delta (Fig. 3A). Intriguingly, these transcripts are upregulated to between one-half and one-third the levels seen for PHB, despite equivalent induction of PHB* and PHB*-Delta (Fig. 3A). As PHB variants are not differentially stable (Supplemental Figs. S1, B to E and S3E), and ChIP experiments indicate near equivalent promoter occupancy (Fig. 2D), these data suggest deletion of the START domain, in addition to reducing the frequency of dimers, significantly reduces their transcriptional potency. This idea is supported by two orthogonal lines of evidence. First, virtually identical results were obtained using co-transfection assays in Nicotiana benthamiana, which show PHB-Delta fails to fully activate both endogenous ZPR targets and a pZPR3:3x-NLS-RFP reporter (Supplemental Fig. S4). Second, the phenotypic severity of transformants constitutively overexpressing PHB-Delta (p35S:PHB*-Delta) is markedly lower than the p35S:PHB* control (Supplemental Fig. S5A). ZPR3 and ZPR4 transcripts in p35S:PHB*-Delta lines also accumulate to between one-half and one-third the levels seen in p35S:PHB* seedlings, despite equivalent accumulation of PHB* and PHB*-Delta transcripts (Supplemental Fig. S5B). These complementary assays support augmenting of transcriptional potency as an additional HD-ZIPIII START regulatory mechanism.
Figure 3.
The START domain enhances PHB transcriptional potency and this property is transferable to heterologous TFs. A) Relative ZPR3 and ZPR4 transcript levels in 24 h estradiol-induced pOlexA:PHB* and pOlexA:PHB*-Delta seedlings indicate PHB-Delta is a less-potent transcriptional activator than PHB. B) Schematic representation of domain-capture-mimicry constructs detailing insertion of the PHB START domain (green) into ATHB12 (purple) and the design of the HD-ZIPI BS:3xNLS-RFP target reporter (top). RT-qPCR following co-transfection with the HD-ZIPI BS:3xNLS-RFP target reporter (bottom) shows fusion of the wild-type PHB START domain augments ATHB12 transcriptional potency by a factor of three (bottom). AS2 is included as negative control. n = 3 biological replicates. *P ≤ 0.05, **P ≤ 0.01, Student's t-test.
START regulatory properties are transferable onto heterologous TFs
The idea that the PHB START domain potentiates TF activity and is not strictly required for TF identity per se is in line with principles of TF evolution by domain capture (Lynch and Wagner 2008; de Mendoza et al. 2013; Jarvela and Hinman 2015; Sebé-Pedrós et al. 2017). Relevant to this, plant genomes encode HD-ZIP TFs that lack a START domain (Ariel et al. 2007; Goodstein et al. 2012; Romani et al. 2018). These related HD-ZIP members provide a unique opportunity to definitively test how addition of a START domain impacts TF output. We therefore inserted the PHB START domain downstream of the homeodomain and leucine zipper motifs of the HD-ZIPI family member ATHB12, partially recapitulating HD-ZIPIII architecture (ATHB12-START). ATHB12 is a known activator of transcription and is sufficient to drive expression of reporters placed downstream of multimeric HD-ZIPI binding sites (Fig. 3B, Lee et al. 2001). Remarkably, addition of the HD-ZIPIII START domain increases ATHB12 transcription activity by a factor of three over unmodified ATHB12 (Fig. 3B). This effect on transcriptional potency parallels effects seen for PHB and PHB-Delta in Arabidopsis as well as equivalent co-transfection assays in N. benthamiana (Figs. 3A and Supplemental Fig. S4). Thus, the START domain is necessary for HD-ZIPIII dimers to achieve full transcriptional potency, and its addition is sufficient to confer this increase in potency onto heterologous TFs.
The START domain does not affect heteromeric stoichiometry
At a molecular level, StARkin domains undergo stereotypical conformational changes in response to ligand binding (Tsujishita and Hurley 2000; Roderick et al. 2002). In the apo form, StARkin domains reveal an open ligand-binding pocket, and upon ligand interaction, the pocket is sealed via conformational changes. In addition to changes in tertiary structure, this creates a new interaction surface that, for a subset of StARkin proteins, mediates recruitment of additional protein partners to exert StARkin-dependent regulatory effects Fujii et al. 2009; Yin et al. 2009; Hubbard et al. 2010; Gatta et al. 2015; Prashek et al. 2017; Tillman et al. 2020; Sanchez-Solana et al. 2021). To test this possibility, we performed quantitative mass spectrometry on proteins that co-immunoprecipitate with PHB (IP-MS; Supplemental Fig. S6). Among the proteins significantly enriched across five PHB IP-MS replicates were multiple members of the BRAHMA-containing Switch/Sucrose Non-fermentable (SWI/SNF) chromatin-remodeling complex. These data point to specificity of the IP-MS and suggest HD-ZIPIII proteins facilitate transcription via direct modification of chromatin. Further supporting specificity of the IP-MS, the known HD-ZIPIII interacting partners ZPR1 and ZPR3 (Wenkel et al. 2007; Kim et al. 2008; Husbands et al. 2016) also co-immunoprecipitate with PHB. Finally, significant enrichments were detected for the lipid-binding proteins OLEOSIN1 (OLEO1), OLEO5, DYNAMIN-RELATED PROTEIN 1C (DRP1C), DRP1E, and ANNEXIN 4 (ANNAT4), which is of potential interest given the lipid-binding nature of StARkin domains (Supplemental Fig. S6 and Supplemental Data Set 1). Protein-protein interactions are, however, not mediated via the START domain as IP-MS shows that PHB-Delta binds the same interaction partners (Supplemental Data Set 2), and SiMPull co-localization analyses confirm that interaction with ZPR3 is unchanged (Supplemental Fig. S7). These findings thus argue against START-mediated regulation of HD-ZIPIII complex stoichiometry at the level of interaction partners.
Taken together, complementary phenotypic and molecular analyses reveal the START domain is required for PHB developmental function, but unlike its homolog in the HD-ZIPIV TFs, does not impact protein stability or subcellular localization. Instead, the presence of a START domain in PHB potentiates TF activity by promoting homodimerization and by increasing transcriptional potency. These effects seem to be mediated by interactions between domains of PHB, as the START domain does not appear to determine its spectrum of interaction partners.
Mutating conserved START residues abolishes PHB DNA-binding competence
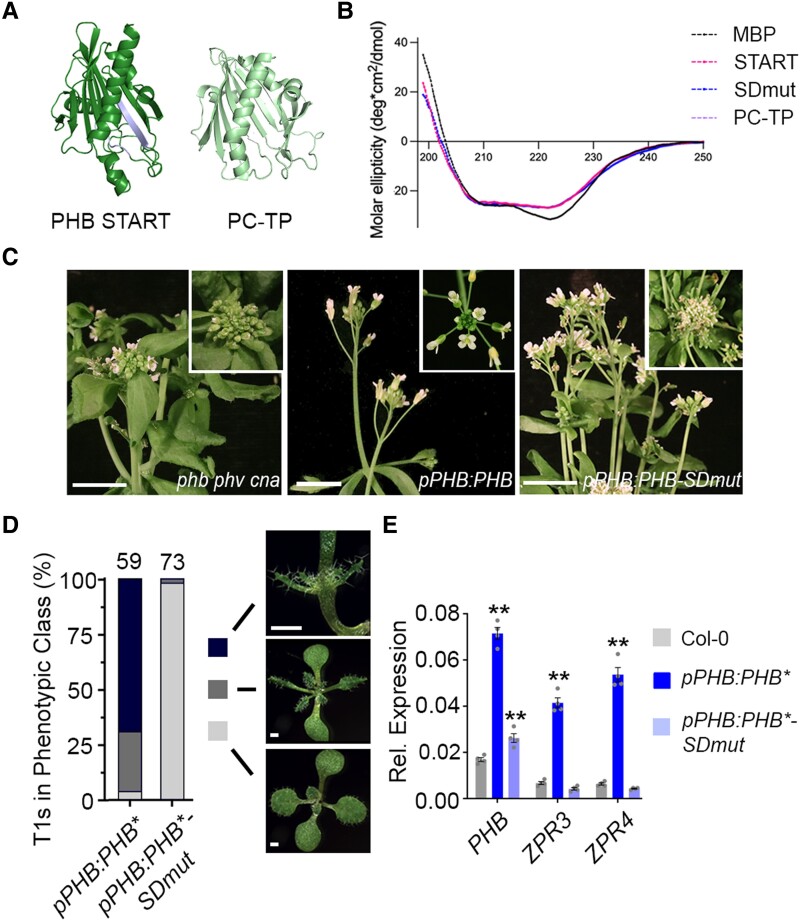
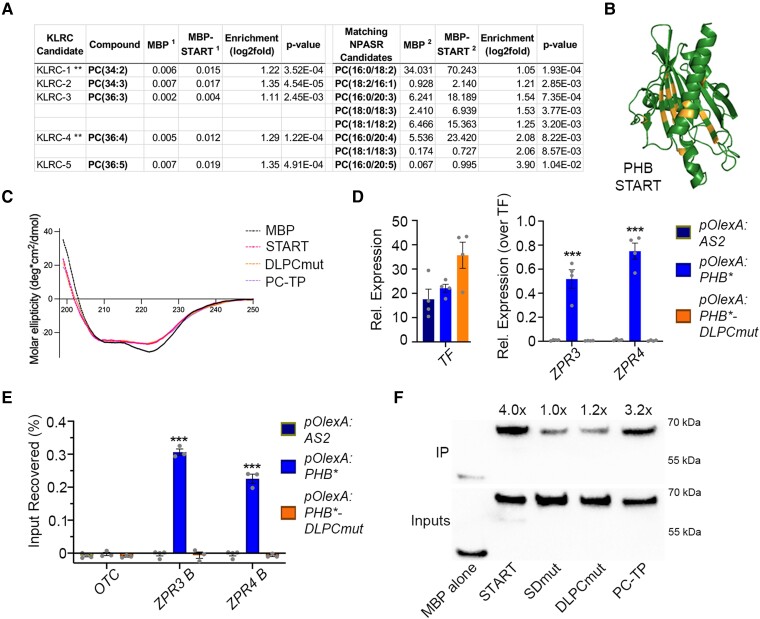
Following the idea that, like other StARkin domains (Prashek et al. 2017; Tillman et al. 2020), the HD-ZIPIII START domain influences intramolecular domain-to-domain interactions, mutations perturbing this property may exert effects on TF function distinct from those observed after full deletion of the domain. To assess this possibility, we first performed homology modeling of the PHB START domain using I-TASSER and AlphaFold2 to identify conserved functional residues to target via mutagenesis. Both algorithms indicate the START domain is distinct from sterol- or isoprenoid-binding StARkin domains such as the abscisic acid (ABA) receptor (Yin et al. 2009), and instead resembles mammalian phosphatidylcholine transfer protein (PC-TP; Fig. 4A, Roderick et al. 2002). START domains like PC-TP contain several highly conserved residues that mediate the ligand-directed conformational change (Ponting and Aravind 1999; Tsujishita and Hurley 2000; Roderick et al. 2002; Baker et al. 2007). Accordingly, two short amino acid stretches (RDFTWLR and RAEMK), centered around three such arginine residues (Ponting and Aravind 1999; Tsujishita and Hurley 2000; Roderick et al. 2002; Baker et al. 2007), were selected for mutagenesis (SDmut; Fig. 4A and Supplemental Fig. S8). Importantly, these mutations are predicted to minimally affect protein folding (RMSD 0.291; Fig. 4A), and circular dichroism confirmed that wild-type, SDmut, and PC-TP START domains purified from E. coli adopt virtually identical secondary structures (Fig. 4B).
Figure 4.
Mutating the START domain perturbs PHB developmental function. A) Homology-guided modeling predicts the PHB START domain (left) closely resembles PC-TP (right). Positions of amino acids mutated in PHB-SDmut are shown in lavender. B) Circular dichroism of MBP (negative control), MBP-START, MBP-SDmut, and MBP-PC-TP shows all three recombinant START proteins have virtually identical secondary structures. C) The pPHB:PHB transgene complements the phb phv cna triple mutant (left vs middle) while pPHB:PHB-SDmut does not (right). D) Phenotypic scoring of primary transformants (n above bar) carrying miR166-insensitive pPHB:PHB* or pPHB:PHB*-SDmut constructs. Ectopic accumulation of PHB leads to severe (black) or intermediate (dark grey) gain-of-function phenotypes, whereas plants mis-accumulating PHB-SDmut appear wild-type (light grey). E) Ectopic PHB* expression leads to strong upregulation of ZPR3 and ZPR4 targets. By contrast, ZPR3 or ZPR4 levels are indistinguishable from Col-0 in pPHB:PHB*-SDmut seedlings, despite accumulation of this variant above endogenous PHB levels. n = 3 biological replicates. **P ≤ 0.01, Student's t-test. Note: PHB, PHB-SDmut, and PHB-Delta data were collected simultaneously. PHB data from Fig. 1 are replotted for clarity.
Identical mutations were then introduced into the functional YFP-tagged pPHB:PHB reporter and its miR166-insensitive counterpart pPHB:PHB*, creating pPHB:PHB-SDmut and pPHB:PHB*-SDmut, respectively. The pPHB:PHB-SDmut transgene failed to complement the phb phv cna triple mutant phenotype (Fig. 4C), suggesting the selected residues are indeed required for PHB function. Supporting this, pPHB:PHB*-SDmut primary transformants did not show phenotypes in the more-sensitive dose-dependent, gain-of-function assay (Fig. 4D), and ZPR3 and ZPR4 transcript levels in these lines were indistinguishable from the wild-type (Fig. 4E). These effects are again not explained by changes in protein stability, sub-cellular localization, or interacting partners, as these properties are comparable for PHB, PHB-Delta, and PHB-SDmut (Supplemental Figs. S1, B to E, S3, S7, and Supplemental Data Set 2). By contrast, SiMPull showed that homodimeric PHB-SDmut complexes are present at ∼40% frequency in the population (Fig. 5A), approximately twice that seen for PHB-Delta but half that seen for wild-type PHB (Fig. 2B).
Figure 5.
Mutating PHB START reduces homodimerization and abolishes DNA-binding competence. A) Percentages of two-step photobleaching events from SiMPull translate to dimerization frequencies of ∼80% for PHB and ∼40% for PHB-SDmut. B) PHB-SDmut does not bind ZPR3 or ZPR4 regulatory regions occupied by PHB. C) Unlike PHB, PHB-SDmut cannot activate ZPR3 or ZPR4 targets in 24 h estradiol-induction experiments. D) Domain-capture-mimicry experiments demonstrate fusion of SDmut START to ATHB12 (ATHB-SDmut) renders ATHB12 non-functional, as RFP reporter levels are indistinguishable from the AS2 negative control. n = 3 biological replicates. **P ≤ 0.01, Student's t-test. Note: PHB, PHB-SDmut, and PHB-Delta data were collected simultaneously. PHB data from Figs. 2 and 3 are replotted for clarity.
We then tested whether PHB-SDmut dimers retain DNA-binding capability. Interestingly, no enrichment of PHB-SDmut was detected in the regulatory regions of the ZPR3 or ZPR4 loci using ChIP (Fig. 5B), indicating these mutations abolish PHB DNA-binding competence. Consistent with this, PHB-SDmut fails to activate ZPR3 and ZPR4 in short-term estradiol-induction assays in Arabidopsis as well as in co-transfection assays in N. benthamiana (Figs. 5C and Supplemental Fig. S4). Moreover, addition of this non-functional START domain to ATHB12 abolishes its transcriptional activity (ATHB12-SDmut; Fig. 5D). Thus, mutating the START domain, and deleting the START domain entirely, leads to proteins with distinct biochemical properties: PHB-SDmut dimerizes relatively well but is unable to bind DNA, whereas PHB-Delta rarely dimerizes but those dimers that do form retain DNA-binding competence and have reduced transcriptional potency.
The PHB START domain binds PC and mutating PC-binding residues abolishes DNA-binding
Structural modeling and mutational analyses propose the PHB START domain may be controlled by phospholipid ligands (Figs. 4A and 5, and Supplemental Fig. S8). We therefore tested whether the PHB START domain interacts with a set of ligands similar to those bound by PC-TP. Depending on context, these include members of the PC, phosphatidylethanolamine (PE), and phosphatidylglycerol (PG) phospholipid classes (de Brouwer et al. 2001; Sablin et al. 2009; Zhai et al. 2017). To this end, recombinant PHB START domain protein was incubated with liposomes generated from Arabidopsis total lipid extracts, re-purified by affinity chromatography, and then subjected to LC-MS at two independent lipidomics centers. Prior to incubation with plant-derived liposomes, recombinant PHB START protein co-purified with bacterial PE and PG species (Supplemental Figs. S9, A and B, Supplemental Data Set 3). Binding of these “fortuitous ligands” has also been reported for other recombinant PC-binding proteins including PC-TP (de Brouwer et al. 2001), steroidogenic factor 1 (SF-1, Sablin et al. 2009), and liver receptor homolog-1 (LRH-1, Zhai et al. 2017). After incubation with plant-derived liposomes (Supplemental Fig. S9C), additional lipids were significantly enriched across five PHB START LC-MS replicates (Supplemental Data Sets 4 to 6). These include five species of PC, two of which are preferred ligands for PC-TP (Fig. 6A; de Brouwer et al. 2001). Further, membrane-overlay assays indicate this binding is not occurring at the surface of the START domain (Supplemental Fig. S9D), drawing further parallels between the HD-ZIPIII and PC-TP START domains.
Figure 6.
PHB START domain binds PC and mutating predicted PC-contacting residues abolishes PC-binding and PHB DNA-binding competence. A) LC-MS analysis of lipids bound by recombinant MBP and MBP-START proteins after liposome incubation and repurification. Several species of PC are significantly enriched in MBP-START. Raw signal intensities are reported, ** indicates species preferentially bound by PC-TP (de Brouwer et al. 2001). B) Homology modeling of DLPCmut START variant showing position of mutations (orange). C) Circular dichroism of MBP, MBP-START, MBP-DLPCmut, and MBP-PC-TP showing all three recombinant START proteins have virtually identical secondary structures. D) Unlike PHB, PHB-DLPCmut cannot activate ZPR3 or ZPR4 targets in 24 h estradiol-induction experiments. E) PHB-DLPCmut does not bind ZPR3 or ZPR4 regulatory regions occupied by PHB. F) MBP fusions of PHB-START and PC-TP strongly bind to PC-conjugated beads, whereas binding of MBP-SDmut and MBP-DLPCmut is indistinguishable from that of the MBP negative control. IP enrichments were calculated by first normalizing each protein to its input then to the MBP alone IP/Input value. n = 3 or 4 biological replicates. ***P ≤ 0.001, Student's t-test.
We therefore created a new PHB START variant with mutations in 17 residues analogous to those contacting PC within the PC-TP binding pocket (DLPCmut; Supplemental Figs. S8 and 9E, Roderick et al. 2002). The substitutions chosen have minimal effects on overall structure (RMSD 0.372; Fig. 6B), and include six amino acids with bulkier side chains that partially occlude the ligand-binding pocket (Supplemental Figs. S9F to G). To test the impact of these mutations, we performed PC-binding assays alongside wild-type, SDmut, and PC-TP START domains after first confirming that all purified START domains have nearly indistinguishable secondary structures (Figs. 4B and 6C). Wild-type and PC-TP START domains both showed strong binding to PC-conjugated beads (Fig. 6F). By contrast, binding affinity of SDmut START domain was indistinguishable from the MBP negative control, indicating that, in addition to their predicted effects on START conformational change, these substitutions abolish PC binding. Loss of PC-binding capacity was also observed with DLPCmut recombinant START protein (Fig. 6F), and although PHB proteins containing the DLPCmut START domain variant (PHB-DLPCmut) are nuclear-localized in plants, these fail to activate ZPR3 and ZPR4 (Fig. 6D; Supplemental Fig. S9H). Moreover, no enrichment of PHB-DLPCmut was detected at these loci using ChIP assays (Fig. 6E). Taken together, these data propose species of PC as promising candidate ligands for the HD-ZIPIII START domain and show that mutations in amino acids that affect ligand binding and possibly ligand-mediated conformational change result in loss of HD-ZIPIII DNA-binding competence (Figs. 4B and 6E).
Discussion
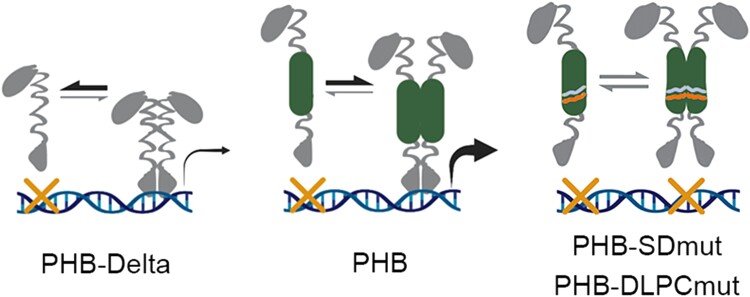
HD-ZIPIII TFs are principal regulators of key developmental innovations throughout plant evolution. Although molecularly cloned over 20 years ago (McConnell et al. 2001), the contribution of their START domain remained elusive. We show that the START domain promotes TF homodimerization and increases transcriptional potency. These effects are mediated solely through intramolecular protein changes and can be ported onto other TFs. These findings are particularly intriguing when considered through the lens of TF evolution by domain capture. Basic HD-ZIP proteins and minimal START proteins resembling PC-TP are both present in unicellular algae (Goodstein et al. 2012; Romani et al. 2018), whereas HD-ZIPIII architecture arose after the divergence of Chlorophyta, but before the emergence of Chlorokybus atmophyticus over 725 million years ago (Wang et al. 2020). Capture of a START domain by HD-ZIPIII antecedents may thus have augmented their transcriptional output. Presumably this also placed their activity under control of a ligand such as PC (Fig. 6A). This supposition is consistent with our data. We find that perturbing START domain residues that affect ligand binding and likely the downstream conformational change abolish DNA-binding competence (Figs. 5B and 6E, Ponting and Aravind 1999; Tsujishita and Hurley 2000; Roderick et al. 2002; Baker et al. 2007). One potential explanation is that the apo form of the START domain holds PHB in a non-DNA-binding conformation. Binding of its ligand, or deletion of the START domain entirely, would relieve this inhibition and permit DNA binding. Taken together, these data, as well as the known properties of StARkin domains (Tsujishita and Hurley 2000; Roderick et al. 2002; Santiago et al. 2009; Hubbard et al. 2010; Tillman et al. 2020; Dresden et al. 2021), present a model in which the START domain potentiates HD-ZIPIII TF activity, and uses ligand-induced conformational change to render HD-ZIPIII dimers competent to bind DNA (Fig. 7).
Figure 7.
The START domain potentiates HD-ZIPIII transcriptional activity. Without a START domain, HD-ZIPIII TFs make fewer and less transcriptionally potent dimers (left), while mutations that prevent ligand binding at the START domain abolish DNA binding (right). START mutations marked (right). These data propose a model in which a functional START domain promotes HD-ZIPIII dimerization and transcriptional potency but requires a ligand for these TFs to bind DNA (middle).
Given this model, it is intriguing to speculate whether acquisition of a START domain enabled HD-ZIPIII TFs to more effectively integrate signaling inputs into their gene networks, a critical feature of multicellularity (Mikhailov et al. 2009; Sebé-Pedrós et al. 2017). In addition, as phospholipid accumulation is spatially regulated (Okazaki and Saito 2014; Stanislas et al. 2018), acquisition of the START domain could have allowed HD-ZIPIII activity to become patterned across groups of cells. This would have clear developmental implications as transcripts of ancestral HD-ZIPIII genes are not targeted by small RNAs of the miR166 family (Floyd et al. 2006). Such a regulatory paradigm draws parallels between HD-ZIPIII TFs and mammalian nuclear receptors (Sladek 2011; Evans and Mangelsdorf 2014); however, future experiments are needed to determine whether START ligands play structural roles or indeed contribute to the intricate spatiotemporal regulation of HD-ZIPIII activity. Either way, the properties of the START domain described here suggest a compelling basis for the emergence of HD-ZIPIII TFs as key drivers of plant morphogenic evolution.
The START domain of the closely related HD-ZIPIV family is similarly required for function, and impacts subcellular localization and protein stability, possibly through binding of epidermally synthesized ceramides (Schrick et al. 2014; Iida et al. 2019; Nagata et al. 2021). The regulatory properties conferred by the HD-ZIPIII START domain are thus distinct, despite their evolutionary relationship (Ponting and Aravind 1999; Schrick et al. 2004). Further, the modulation of DNA-binding competence identified here represents a new type of StARkin-directed regulatory mechanism. Our findings, in addition to resolving a long-standing mystery in plant development, highlight the flexible and diverse regulatory potential coded within this widely distributed evolutionary module (Dresden et al. 2021).
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana (Col-0 ecotype) seedlings and Nicotiana benthamiana plants were grown at 22 °C under long-day conditions, on soil or 1% agarose plates containing Murashige and Skoog medium supplemented with 1% sucrose. Light bulbs were Phillips (F32T8/TL965/ALTO TG 30PK) with a Kelvin temperature of 6500 K and lumens of 2600. Inductions were performed by spraying 10-day-old seedlings with 20 uM B-estradiol in 1% DMSO supplemented with 0.005% Silwet.
Molecular biology and plant transformations
pPHB:PHB-YFP and pPHB:PHB*-YFP constructs have been described previously (Skopelitis et al. 2017). Using these templates, pPHB:PHB-Delta-YFP and pPHB:PHB*-Delta-YFP were constructed via Gibson assembly (NEB), replacing the 642 bp START domain (496 to 1137 bp from the start codon) with a sensitive (GGGATGAAGCCTGGTCCGGAT) or an insensitive version (GGGATGAAGCCTGGACCGGAT) of the 21nt miR166-recognition site (Skopelitis et al. 2017). To create pPHB:PHB-SDmut-YFP, we first synthesized a mutated variant of the START domain (Mr. Gene), which replaced amino acids RDFTWLR with GAVVGAG and amino acids RAEMK with VAAGV by including the following nucleotide substitutions: CGTGACTTTTGGACGCTGAGA at position 841 to 861 bp from the start codon to GGTGCCGTCGTAGGAGCAGGC, and AGAGCTGAAATGAAA at position 961 to 975 bp from the start codon to GTGGCGGCCGGCGTC. miR166-insensitive pPHB:PHB*-SDmut-YFP was then constructed via site-directed mutagenesis (Stratagene), copying the mutations in pPHB:PHB*-YFP (Skopelitis et al. 2017). All constructs were shuttled into the pB7GW binary vector via Gateway LR reactions (VIB Ghent; Invitrogen).
To facilitate subsequent stable and inducible overexpression in Arabidopsis, and transient expression in tobacco, PHB*-YFP, PHB*-SDmut-YFP, and PHB*-Delta-YFP cDNAs were reamplified and cloned into pCR8-GW via Gibson assembly (Invitrogen; NEB). Gateway LR reactions then shuttled each cDNA into pEARLYGATE100 (stable overexpression), modified pMDC7 with the UBQ10 promoter in place of G10-90 (inducible overexpression), or p502Ω (transient expression; VIB Ghent). PHB-DLPCmut*-YFP cDNA was generated by replacing the wild-type START domain with a gene-synthesized mutant variant (GeneArt) using Gibson cloning (ThermoFisher). A Gateway LR reaction then shuttled PHB-DLPCmut-YFP into pMDC7. ATHB12 was amplified from cDNA generated from seedlings, then cloned into pCR8-GW using Gibson (NEB). Fusions between ATHB12 and the START domain variants were created by inserting a fragment of PHB* or PHB-SDmut (376 to 1137 bp from the start codon) into ATHB12 (at position 366 from the start codon) using Gibson assembly (Invitrogen; NEB). Gateway LR reactions then shuttled these cDNAs into p502Ω (Invitrogen; VIB Ghent).
The pZPR3:3xNLS-RFP reporter was created by first cloning the 3.2 kb region upstream of the ZPR3 start codon into pCR8-GW (Invitrogen; NEB), followed by an LR reaction using a modified pGREEN binary vector with a Gateway cassette upstream of 3xNLS-RFP (Invitrogen). The HD-ZIPI BS:3xNLS-RFP reporter was created by first synthesizing a construct comprised of six copies of the HD-ZIPI binding site (CAATTATTG) followed by a minimal 35S enhancer, flanked by attL1/attL2 sites (Mr. Gene). A 10-nt spacer (CATTTCAAGA) was inserted between each binding site to minimize potential stearic hindrance. Finally, an LR reaction was used to shuttle these multimerized binding sites into the pGREEN binary described above (Invitrogen). Cloning primer sequences for all constructs are listed in Supplemental Data Set 7. Synthesized sequences are in Supplemental Data Set 8.
Transient transfection of N. benthamiana was performed using syringe-mediated infiltration (Sparkes et al. 2006). In brief, overnight cultures of Agrobacterium tumefaciens were centrifuged, resuspended in 2 to 5 mL of room temperature infiltration medium (10 mM MgCl2, 10 mM MES KOH, pH 5.6, 150 mM acetosyringone [Sigma Aldrich], and 1% DMSO), diluted to a working optical density of 1, and infiltrated into third and fourth leaves of 3- to 4-week-old N. benthamiana plants.
Homology modeling and sequence alignment
The PHB START domain was modeled with I-TASSER (https://zhanggroup.org/I-TASSER/) and AlphaFold2 (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb) using default parameters. Sequences were aligned using the ClustalW algorithm in the MEGA X software (v.10.1.7; https://www.megasoftware.net/). The crystal structure of PC-TP (1ln1) was the top template used for threading in I-TASSER. Models were visualized in Pymol.
ChIP and RT-qPCR assays
ChIP assays were performed with 11-day-old estradiol-induced seedlings as previously described (Husbands et al. 2015), using IgG (abcam ab46540) or anti-GFP (abcam ab290) antibodies. ChIP and input DNA samples were assayed by qPCR using iQ SYBR Green Supermix (Bio-Rad). ZPR3 and ZPR4 regulatory regions assayed in ChIP were selected based on the presence of HD-ZIPIII binding sites predicted by FIMO (https://meme-suite.org/meme/tools/fimo). All experiments were performed at least three independent times. PCR was performed in duplicate, and enrichments calculated relative to input. Student's t-test was used to calculate statistical significance.
Total RNA was extracted from seedlings or infiltrated N. benthamiana leaves using Trizol reagent (Gibco BRL). One microgram of RNA was primed with oligo (dT) and reverse transcribed using the SuperScript III first-strand synthesis kit (Invitrogen). Relative quantification values were calculated based on at least three biological replicates, with ΔCt of ACT2 or B-tubulin serving as normalization controls in Arabidopsis or N. benthamiana, respectively. Wild-type or uninduced values were set to one and PHB variant values either plotted directly or after further normalization to PHB variant levels. Student's t-test was used to calculate statistical significance. ChIP and RT-qPCR primer sequences are listed in Supplemental Data Set 9.
Plant imaging
Brightfield images of Arabidopsis seedlings were captured using an SMZ1500 dissecting microscope with NIS Element software (Nikon). Fluorescent images of Arabidopsis seedlings and infiltrated N. benthamiana leaves were obtained using the same microscope with the P-FLA2 epi-fluorescent attachment. Heart-stage embryos were dissected, stained with Fluorescent Brightener 28 (Sigma Aldrich), and then imaged using an LSM780 confocal microscope (Zeiss).
Single-molecule imaging and stoichiometric analyses
A detailed protocol of SiMPull with plant tissue has been published (Husbands et al. 2016); however, the amount of input tissue can vary between experiments. Here, lysates for SiMPull were prepared from flash frozen tissue comprised of five-to-six 10-day-old Arabidopsis seedlings or 1 to 2 cm2 pieces of infiltrated N. benthamiana leaves. Note: SiMPull experiments in Arabidopsis were conducted using seedlings expressing PHB variants under native regulatory elements (Figs. 2, A and B) as well as an estradiol-inducible promoter (Supplemental Fig. S3D). A minimum of three independent biological replicates were performed for each SiMPull experiment. To enable calculation of PHB variant dimerization frequencies, as well as the maturation probability of citrine YFP in Arabidopsis, 2 × 35S:monoYFP and 2 × 35S:tdYFP constructs (described in Husbands et al. 2016) were stably transformed and at least four independent lines analyzed.
SiMPull was first performed with 1:150 dilutions of lysates from 2 × 35S:monoYFP or 2 × 35S:tdYFP seedlings, mixed at ratios described in Supplemental Fig. S2. Frequencies of two-step photobleaching events were then scored and used to construct a standard calibration curve (y = 2.28x - 27; R2 = 0.992). For stoichiometric analyses of PHB variants, lysates from native promoter lines or 24 h estradiol-induced lines were diluted 1:1 or 1:100, respectively, then subject to SiMPull and photobleaching counts. Frequencies of two-step photobleaching events for each genotype were translated into dimerization frequencies via the above calibration curve. Maturation probability was predicted using binomial probability modeling (described in Husbands et al. 2016). Co-localization data for ZPR3-mCherry with AS2-YFP, PHB-YFP, PHB-SDmut-YFP, or PHB-Delta-YFP were collected and analyzed as described previously (Husbands et al. 2016).
Quantification of PHB variant induction with estradiol
Ten-to-twelve 10-day-old seedlings were induced for 24 h with estradiol, flash frozen, then ground in 500 µl freshly prepared lysis buffer (25 mM Tris HCl pH 8, 150 mM NaCl, 1% SDS, 1 × cOmplete ULTRA protease inhibitor (Roche), and 1 × PhosSTOP (Roche)). Lysates were cleared via 14,000 × g centrifugation at 4 °C, and split evenly for RNA vs protein processing. RNA extraction, cDNA synthesis, and RT-qPCR were carried out as described above. Lysates for protein work were mixed 1:1 with 2 × Laemmli sample buffer, and boiled for 1 min. Proteins were resolved via SDS-PAGE, blotted to Hybond ECL membrane (GE Healthcare), and blocked in 5% milk fat. PHB variants were detected via anti-GFP primary antibodies (600-406-215; Rockland Immunochemicals; 1:500 dilution) and anti-rabbit horseradish peroxidase-conjugated secondary antibodies (Jackson Immunoresearch; 1:5,000 dilution). Detection of secondary antibodies was performed with SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher Scientific). Blots were scanned and quantified using ImageJ. Four independent biological replicates were performed, each with two technical replicates.
Protein immunoprecipitation and mass spectrometry
One gram of 24 h estradiol-induced seedlings was flash frozen, ground under liquid nitrogen, and extracted in 4 ml of freshly prepared extraction buffer (50 mM HEPES pH 8, 150 mM NaCl, 0.3% NP-40, 1 × cOmplete ULTRA protease inhibitor -EDTA (Roche), 1 mM PMSF, 50 uM bortezomib). Lysates were cleared via 14,000 × g centrifugation at 4 °C, pre-cleared with Protein A Dynabeads, and incubated for 1 h with 5 µl anti-GFP antibody (abcam ab290). PHB variant complexes were captured using 25 µl Protein A Dynabeads and washed 4 × with freshly prepared Wash Buffer (50 mM HEPES pH 8, 450 mM NaCl, 0.1% NP-40, 1 × cOmplete ULTRA protease inhibitor -EDTA (Roche), 1 mM PMSF, 50 µM bortezomib).
After the final wash, beads were flash frozen in liquid nitrogen and trypsinized via standard protocols (Promega). Peptides were labeled with 8-plex iTRAQ (Ross et al. 2004), and injected into the Orbitrap Velos Pro mass spectrometer. Protein identification and quantification was carried using Mascot 2.4 (Perkins et al. 1999) against the UniProt Arabidopsis sequence database. Enrichments over the uninduced control were calculated for each replicate, mean enrichment was then calculated across the top five replicates, and a Student's t-test applied to determine significance. Known false positives were eliminated from subsequent analyses (Van Leene et al. 2015). Interacting partners for PHB-SDmut and PHB-Delta were identified using a similar workflow. Raw data are included in Supplemental Data Set 10.
Protein purification, circular dichroism, membrane-overlay, and PC-conjugated bead assays
Wild-type or START mutant variants (496 to 1137 bp from the start codon) were cloned downstream of maltose binding protein (MBP), in a modified pET28b vector (Laha et al. 2015), via Gibson cloning (ThermoFisher). Proteins were induced with 100 µM IPTG at 12 °C for 16 h and purified on a Ni2+ column according to manufacturer protocols (Qiagen). Proteins were eluted using 125 mM imidazole, desalted and concentrated on a 10 MWCO column (Amersham) into NaCl-free IP buffer (1 × PBS pH7.4, 5% glycerol), and quantified next to BSA on a 10% SDS gel.
Circular dichroism was performed with the Jasco J-815 Spectrometer (OSU Biophysical Interaction & Characterization Facility) using 1 mg/ml of purified MBP or MBP-START variant proteins, then corrected and converted to molar ellipticity in Excel (Microsoft). Scans were limited to a range of 190 to 250 nm to increase resolution and data were plotted in GraphPad (Prism).
Membrane-overlay assays were done according to manufacturer protocols (ThermoFisher Scientific; P23751). In brief, membranes were blocked for 1 h at RT (PBS, 0.1% Tween20, 3% BSA fatty acid free). One hundred fifty micrograms of wild-type START purified protein was added to the membrane in 5 ml blocking buffer and incubated for 1 h with gentle shaking. Membranes were washed three times with PBS-T and incubated with 1:2,000 anti-His-tag antibody (Abiocode M0335-1) for 1 h with gentle shaking at RT. Membranes were washed three times and incubated with 1:2,000 anti-mouse 2o antibody (ab6789) for 1 h. Bound proteins were detected using Hyper HRP Substrate (Takara). PtIns(3,5)P2 Grip protein (P-3516-3-EC, MoBiTec), detected via anti-GST antibody (Sigma Aldrich, A7340), served as a positive control (not shown).
PC-conjugated bead assays were done according to manufacturer protocols (P-B0PC; Echelon Biosciences) except that 0.1% IGEPAL was used in Buffer System 1. In brief, 1 nmol of purified START domain protein was added to 0.25 nmols of beads equilibrated in Buffer System 1. Protein-bead mixes were rotated at room temperature for 15 min, then transferred to 4 °C for continued capture overnight. Protein-bead mixes were washed 3 × using 250 ul Buffer System 1, eluted using 1 × Laemmli buffer, and bound proteins separated on an SDS-PAGE gel. Bound proteins and 1:1,000 diluted Inputs were detected by western blotting using a 1:1,000 HRP-conjugated 6x-His Tag antibody (PA1-23024; ThermoFisher).
Liposome production and dynamic light scattering
Total lipids from 35 to 40 g of 21d old Arabidopsis seedling tissue were extracted using Bligh-Dyer (Bligh and Dyer 1959). Lipids were dried under argon and resuspended in 100% ethanol to 100 mg/ml. Liposomes were generated by rapid injection of ethanol-suspended lipids into IP buffer (1 × PBS pH7.4, 150 mM NaCl, 5% glycerol). Liposome size range was determined by dynamic light scattering using the Zetasizer Nano (Malvern).
Lipid immunoprecipitations and mass spectrometry
To identify lipids bound by the START domain in bacteria, lipids from purified recombinant MBP and MBP-START proteins were immediately extracted using Bligh & Dyer (Bligh and Dyer 1959), dried under argon, and resuspended in chloroform. Lipids were identified using LC-MS performed at the Institut fur Molekulare Physiologie und Biotechnologie der Pflanze (IMBIO) at the University of Bonn, Germany. In brief, lipids were resuspended in 200 µl Q-ToF solvent was added (methanol/chloroform/300 mM ammonium acetate, 665:300:35, v/v/v, Welti et al. 2002), and measured using nanoflow direct infusion Q-TOF MS/MS (as described in Gasulla et al. 2013).
To identify lipids bound by the START domain in plant lipid mixtures, 1 mg of recombinant MBP or MBP-START proteins were incubated with 6 mg of liposomes in IP buffer (1 × PBS pH 7.4, 150 mM NaCl, 5% glycerol) overnight at 4 degrees. IPs were performed in quintuplicate. Proteins were repurified using nickel affinity chromatography, extensively washed, and eluted using 125 mM imidazole. Lipids were extracted using Bligh & Dyer (Bligh and Dyer 1959), dried under argon, and resuspended in chloroform. Lipids were identified using LC-MS performed at the Kansas Lipidomics Research Center Analytical Laboratory (KLRC; Kansas State University) and the Nutrient & Phytochemical Analytics Shared Resource (NPASR; Ohio State Comprehensive Cancer Center). KLRC used a Waters Xevo TQS mass spectrometer adjusted for SPLASH response factors. Four hundred microliter volumes were injected, ES + ionization mode was used for all detection of all compounds except lysoPG which used ES-, and data presented are nmol per mg dry weight and normalized to mols of protein per IP. NPASR data were using a SelexION/QTrap 5500 (Sciex) Lipidyzer for CER, LPC, LPE, PC, PE, and SM and a 6550 QTOF MS (Agilent) for FFAs. One hundred fifty microliter volumes were injected, ESI- ionization mode was used, and data presented are normalized to mols of protein per IP.
We note that KLRC instrument acquisition and lipidomics method development was supported by the National Science Foundation (EPS 0236913, MCB 1413036, MCB 0920663, DBI 0521587, DBI1228622), Kansas Technology Enterprise Corporation, K-IDeA Networks of Biomedical Research Excellence (INBRE) of National Institute of Health (P20GM103418), and Kansas State University. We also note that research reported in this publication was supported by The Ohio State University Comprehensive Cancer Center and the National Institutes of Health under grant number P30 CA016058.
Accession numbers
PHABULOSA – AT2G34710; PHAVOLUTA – AT1G30490; REVOLUTA – AT5G60690; CORONA -AT1G52150; ATHB8 – AT4G32880; LITTLE ZIPPER3 – AT3G52770; LITTLE ZIPPER4 – AT2G36307; ATHB12 – AT3G61890; OLEOSIN1 – AT4G25140; DYNAMIN-RELATED PROTEIN 1C – AT1G14830; DYNAMIN-RELATED PROTEIN 1E – AT3G60190; ANNEXIN 4 – AT2G38750; ORNITHINE TRANSCARBAMYLASE (AT1G75330).
Supplementary Material
Acknowledgments
We are grateful to D. Skopelitis, L. Joshua-Tor, D. Pappin, and K. Rivera from Cold Spring Harbor Laboratory, Diana Vranjkovic and Phillipp Johnen from the University of Tuebingen, and Brian J. Smith at Ohio State University for technical support. We are grateful to Dr. Zachary Schultz at Ohio State University for assistance with dynamic light scattering assays, and Dr. Alicia Friedman at Ohio State University for assistance with circular dichroism. We thank Dr. Ruth Welti and Mary Roth at the KLRC, and Dr. Ken Riedl at the OSUCCC NPASR, for lipidomic analyses. We thank Dr. Mark Stahl at the Center for Plant Molecular Biology at the University of Tübingen for assistance with LC-MS. We also thank Dr. Doris Wagner for critical comments on the manuscript. Support for this work came from the National Science Foundation grants IOS-1022102 and IOS-1355018, as well as the Alexander von Humboldt Professorship, to M.T. Support for this work also came from the National Science Foundation grant IOS-2039489 to A.Y.H.
Contributor Information
Aman Y Husbands, Cold Spring Harbor Laboratory, 1 Bungtown Road, Cold Spring Harbor, NY 11724, USA; Department of Biology, University of Pennsylvania, 415 S. University Ave, Philadelphia, PA 19104, USA.
Antje Feller, Center for Plant Molecular Biology, University of Tübingen, Auf der Morgenstelle 32, 72076 Tübingen, Germany.
Vasudha Aggarwal, Department of Biophysics and Biophysical Chemistry, Johns Hopkins School of Medicine, Baltimore, MD 21205, USA.
Courtney E Dresden, Department of Biology, University of Pennsylvania, 415 S. University Ave, Philadelphia, PA 19104, USA; Molecular, Cellular, and Developmental Biology (MCDB), The Ohio State University, Columbus, OH 43215, USA.
Ashton S Holub, Department of Molecular Genetics, The Ohio State University, Columbus, OH 43215, USA.
Taekjip Ha, Department of Biophysics and Biophysical Chemistry, Johns Hopkins School of Medicine, Baltimore, MD 21205, USA; Department of Biophysics and Biophysical Chemistry, Johns Hopkins School of Medicine, Howard Hughes Medical Institute, Baltimore, MD 21205, USA.
Marja C P Timmermans, Cold Spring Harbor Laboratory, 1 Bungtown Road, Cold Spring Harbor, NY 11724, USA; Center for Plant Molecular Biology, University of Tübingen, Auf der Morgenstelle 32, 72076 Tübingen, Germany.
Author contributions
A.Y.H and M.T conceived of the research direction and experiments; A.Y.H. performed experiments except: protein purification by A.F., LC-MS by KLRC and NPASR centers, additional ChIP and protein work by C.E.D. and A.S.H, PC-binding assays by C.E.D., and SiMPull by V.A. under the guidance of T.H. A.Y.H and M.T. wrote the manuscript.
Supplemental data
Supplemental Figure S1 . The disparate behaviors of PHB variants are not explained by differences in transcript accumulation, nuclear protein localization, or ectopic expression in the absence of miR166-regulation.
Supplemental Figure S2 . Citrine YFP maturation probability in Arabidopsis and construction of a SiMPull calibration curve.
Supplemental Figure S3 . Subcellular localization, dimerization frequency, and protein stability are unaffected upon estradiol-induced overexpression of PHB variants.
Supplemental Figure S4 . The START domain increases transcriptional potency.
Supplemental Figure S5 . PHB-Delta dimers have reduced transcriptional potency.
Supplemental Figure S6 . PHB binds multiple classes of interacting partners in a START-independent manner.
Supplemental Figure S7 . PHB—ZPR3 interaction is not affected by START mutation or deletion.
Supplemental Figure S8 . The PHB START domain most closely resembles PC-TP.
Supplemental Figure S9 . The START domain binds phospholipids, and mutations abolishing PC binding do not affect folding or nuclear localization.
Supplemental Data Set 1 . Proteins co-immunoprecipitating with PHB.
Supplemental Data Set 2 . Mutation or deletion of the START domain does not alter the suite of PHB-interacting partners.
Supplemental Data Set 3 . PE and PG species co-purified from bacteria at IMBIO (Bonn).
Supplemental Data Set 4 . Compounds identified at KLRC after incubation with Arabidopsis liposomes and subsequent repurification.
Supplemental Data Set 5 . Compounds identified at NPASR facility after incubation with Arabidopsis liposomes and subsequent repurification.
Supplemental Data Set 6 . Compounds significantly enriched in both KLRC and NPASR data sets (Arabidopsis liposomes).
Supplemental Data Set 7 . Cloning primers.
Supplemental Data Set 8 . Synthesized sequences.
Supplemental Data Set 9 . ChIP and RT-qPCR primers.
Supplemental Data Set 10 . Raw data used to generate Supplemental Data Set 1 (Proteins co-immunoprecipitating with PHB).
Data availability
All study data are included in the article or the supporting information.
References
- Aggarwal V, Ha T. Single-molecule pull-down (SiMPull) for new-age biochemistry: methodology and biochemical applications of single-molecule pull-down (SiMPull) for probing biomolecular interactions in crude cell extracts. BioEssays. 2014:36(11):1109–1119. 10.1002/bies.201400090 [DOI] [PubMed] [Google Scholar]
- Alpy F, Tomasetto C. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J Cell Sci. 2005:118(13):2791–2801. 10.1242/jcs.02485 [DOI] [PubMed] [Google Scholar]
- Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-Zip family. Trends Plant Sci. 2007:12(9):419–426. 10.1016/j.tplants.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Baker BY, Epand RF, Epand RM, Miller WL. Cholesterol binding does not predict activity of the steroidogenic acute regulatory protein, StAR. J Biol Chem. 2007:282(14):10223–10232. 10.1074/jbc.M611221200 [DOI] [PubMed] [Google Scholar]
- Belda-Palazon B, Gonzalez-Garcia MP, Lozano-Juste J, Coego A, Antoni R, Julian J, Peirats-Llobet M, Rodriguez L, Berbel A, Dietrich D, et al. . PYL8 mediates ABA perception in the root through non-cell-autonomous and ligand-stabilization-based mechanisms. Proc Natal Acad Sci U S A. 2018:115(50):E11857–E11863. 10.1073/pnas.1815410115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959:37(1):911–917. 10.1139/y59-099 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. . Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010:465(7296):316–321. 10.1038/nature08977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brouwer AP, Bouma B, van Tiel CM, Heerma W, Brouwers JF, Bevers LE, Westerman J, Roelofsen B, Wirtz KW. The binding of phosphatidylcholine to the phosphatidylcholine transfer protein: affinity and role in folding. Chem Phy Lipids. 2001:112(2):109–119. 10.1016/S0009-3084(01)00171-2 [DOI] [PubMed] [Google Scholar]
- de Mendoza A, et al. . Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc Natl Acad Sci USA. 2013:110(50):E4858–E4866. 10.1073/pnas.1311818110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresden CE, Ashraf Q, Husbands AY. Diverse regulatory mechanisms of StARkin domains in land plants and mammals. Curr Opin Plant Biol. 2021:64:102148. 10.1016/j.pbi.2021.102148 [DOI] [PubMed] [Google Scholar]
- Du X, Qian X, Papageorge A, Schetter AJ, Vass WC, Liu X, Braverman R, Robles AI, Lowy DR. Functional interaction of tumor suppressor DLC1 and caveolin-1 in cancer cells. Cancer Res. 2012:72(17):4405–4416. 10.1158/0008-5472.CAN-12-0777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM, Mangelsdorf DJ. Nuclear receptors, RXR, and the big bang. Cell. 2014:157(1):255–266. 10.1016/j.cell.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd SK, Zalewski CS, Bowman JL. Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics. 2006:173(1):373–388. 10.1534/genetics.105.054239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009:462(7273):660–664. 10.1038/nature08599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasulla F, Vom Dorp K, Dombrink I, Zähringer U, Gisch N, Dörmann P, Bartels D. The role of lipid metabolism in the acquisition of desiccation tolerance in Craterostigma plantagineum: a comparative approach. Plant J. 2013:75(5):726–741. 10.1111/tpj.12241 [DOI] [PubMed] [Google Scholar]
- Gatta AT, Wong LH, Sere YY, Calderón-Noreña DM, Cockcroft S, Menon AK, Levine TP. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. eLife. 2015:4: e07253. 10.7554/eLife.07253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. . Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012:40(D1): D1178–D1186. 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 2010:24(16):1695–1708. 10.1101/gad.1953910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands AY, Aggarwal V, Ha T, Timmermans MC. In planta single-molecule pull-down reveals tetrameric stoichiometry of HD-ZIPIII:LITTLE ZIPPER complexes. Plant Cell. 2016:28(8):1783–1794. 10.1105/tpc.16.00289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands AY, Benkovics AH, Nogueira FT, Lodha M, Timmermans MC. The ASYMMETRIC LEAVES complex employs multiple modes of regulation to affect adaxial-abaxial patterning and leaf complexity. Plant Cell. 2015:27(12):3321–3335. 10.1105/tpc.15.00454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida H, Yoshida A, Takada S. ATML1 activity is restricted to the outermost cells of the embryo through post-transcriptional repressions. Development. 2019:146(4):dev169300. 10.1242/dev.169300 [DOI] [PubMed] [Google Scholar]
- Jain A, Arauz E, Aggarwal V, Ikon N, Chen J, Ha T. Stoichiometry and assembly of mTOR complexes revealed by single-molecule pulldown. Proc Natal Acad Sci U S A. 2014:111(50):17833–17838. 10.1073/pnas.1419425111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Liu R, Ramani B, Arauz E, Ishitsuka Y, Ragunathan K, Park J, Chen J, Xiang YK, Ha T. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011:473(7348):484–488. 10.1038/nature10016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvela MC, Hinman VG. Evolution of transcription factor function as a mechanism for changing metazoan developmental gene regulatory networks. Evodevo. 2015:6(1):3. 10.1186/2041-9139-6-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004:428(6978):84–88. 10.1038/nature02363 [DOI] [PubMed] [Google Scholar]
- Kanno K, Wu MK, Agate DS, Fanelli BJ, Wagle N, Scapa EF, Ukomadu C, Cohen DE. Interacting proteins dictate function of the minimal START domain phosphatidylcholine transfer protein/StarD2. J Biol Chem. 2007:282(42):30728–30736. 10.1074/jbc.M703745200 [DOI] [PubMed] [Google Scholar]
- Kelley DR, Skinner DJ, Gasser CS. Roles of polarity determinants in ovule development. Plant J. 2009:57(6):1054–1064. 10.1111/j.1365-313X.2008.03752.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Lee M, Lee I, Park HY, Seo PJ, Jung JH, Kwon EJ, Suh SW, Paek KH, et al. . HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell. 2008:20(4):920–933. 10.1105/tpc.107.057448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laha D, Johnen P, Azevedo C, Dynowski M, Weiß M, Capolicchio S, Mao H, Iven T, Steenbergen M, Freyer M, et al. . VIH2 regulates the synthesis of inositol pyrophosphate InsP8 and jasmonate-dependent defenses in Arabidopsis. Plant cell. 2015:27(4):1082–1097. 10.1105/tpc.114.135160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Oh HS, Cheon CI, Hwang IT, Kim YJ, Chun JY. Structure and expression of the Arabidopsis thaliana homeobox gene Athb-12. Biochem Biophys Res Commun. 2001:284(1):133–141. 10.1006/bbrc.2001.4904 [DOI] [PubMed] [Google Scholar]
- Lumba S, Cutler S, McCourt P. Plant nuclear hormone receptors: a role for small molecules in protein-protein interactions. Annu Rev Cell Dev Biol. 2010:26(1): 445–469. 10.1146/annurev-cellbio-100109-103956 [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Wagner GP. Resurrecting the role of transcription factor change in developmental evolution. Evolution. 2008:9(9): 2131–2154. 10.1111/j.1558-5646.2008.00440.x [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5' Region. EMBO J. 2004:23(16):3356–3364. 10.1038/sj.emboj.7600340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001:411(6838):709–713. 10.1038/35079635 [DOI] [PubMed] [Google Scholar]
- Mikhailov KV, Konstantinova AV, Nikitin MA, Troshin PV, Rusin LY, Lyubetsky VA, Panchin YV, Mylnikov AP, Moroz LL, Kumar S, et al. . The origin of metazoa: a transition from temporal to spatial cell differentiation. BioEssays. 2009:31(7):758–768. 10.1002/bies.200800214 [DOI] [PubMed] [Google Scholar]
- Nagata K, Ishikawa T, Kawai-Yamada M, Takahashi T, Abe M. Ceramides mediate positional signals in Arabidopsis thaliana protoderm differentiation. Development. 2021:148(2):dev194969. 10.1242/dev.194969 [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Saito K. Roles of lipids as signaling molecules and mitigators during stress response in plants. Plant J. 2014:79(4):584–596. 10.1111/tpj.12556 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. . Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009:324(5930):1068–1071. 10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999:20(18):3551–3567. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci. 1999:24(4):130–132. 10.1016/S0968-0004(99)01362-6 [DOI] [PubMed] [Google Scholar]
- Prashek J, Bouyain S, Fu M, Li Y, Berkes D, Yao X. Interaction between the PH and START domains of ceramide transfer protein competes with phosphatidylinositol 4-phosphate binding by the PH domain. J biol Chem. 2017:292(34):14217–14228. 10.1074/jbc.M117.780007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005:17(1):61–76. 10.1105/tpc.104.026161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P, Carlsbecker A, Etchells JP. Class III HD-ZIPs govern vascular cell fate: an HD view on patterning and differentiation. J Exp Bot. 2017:68(1):55–69. 10.1093/jxb/erw370 [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002:110(4):513–520. 10.1016/S0092-8674(02)00863-2 [DOI] [PubMed] [Google Scholar]
- Robischon M, Du J, Miura E, Groover A. The Populus class III HD ZIP, popREVOLUTA, influences cambium initiation and patterning of woody stems. Plant Physiol. 2011:155(3):1214–1225. 10.1104/pp.110.167007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderick SL, Chan WW, Agate DS, Olsen LR, Vetting MW, Rajashankar KR, Cohen DE. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat Struct Biol. 2002:9(7):507–511. 10.1038/nsb812 [DOI] [PubMed] [Google Scholar]
- Romani F, Reinheimer R, Florent SN, Bowman JL, Moreno JE. Evolutionary history of HOMEODOMAIN LEUCINE ZIPPER transcription factors during plant transition to land. New Phytol. 2018:219(1):408–421. 10.1111/nph.15133 [DOI] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. . Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004:3(12):1154–1169. 10.1074/mcp.M400129-MCP200 [DOI] [PubMed] [Google Scholar]
- Sablin EP, Blind RD, Krylova IN, Ingraham JG, Cai F, Williams JD, Fletterick RJ, Ingraham HA. Structure of SF-1 bound by different phospholipids: evidence for regulatory ligands. Mol Endocrinol. 2009:23(1):25–34. 10.1210/me.2007-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Solana B, Wang D, Qian X, Velayoudame P, Simanshu DK, Acharya JK, Lowy DR. The tumor suppressor activity of DLC1 requires the interaction of its START domain with Phosphatidylserine, PLCD1, and Caveolin-1. Mol Cancer. 2021:20(1):141. 10.1186/s12943-021-01439-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Márquez JA. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009:462(7273):665–668. 10.1038/nature08591 [DOI] [PubMed] [Google Scholar]
- Schrick K, Bruno M, Khosla A, Cox PN, Marlatt SA, Roque RA, Nguyen HC, He C, Snyder MP, Singh D, et al. . Shared functions of plant and mammalian StAR-related lipid transfer (START) domains in modulating transcription factor activity. BMC Biol. 2014:12(1):70. 10.1186/s12915-014-0070-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick K, Nguyen D, Karlowski WM, Mayer KF. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 2004:5(6):R41. 10.1186/gb-2004-5-6-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian J, Ryu KH, Zhou J, Tarkowská D, Tarkowski P, Cho YH, Yoo SD, Kim ES, Lee JY. PHABULOSA controls the quiescent center-independent root meristem activities in Arabidopsis thaliana. PLoS Genet. 2015:11(3):e1004973. 10.1371/journal.pgen.1004973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Degnan BM, Ruiz-Trillo I. The origin of metazoa: a unicellular perspective. Nat Rev Genet. 2017:18(8):498–512. 10.1038/nrg.2017.21 [DOI] [PubMed] [Google Scholar]
- Sessa G, Steindler C, Morelli G, Ruberti I. The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol Biol. 1998:38(4):609–622. 10.1023/A:1006016319613 [DOI] [PubMed] [Google Scholar]
- Skopelitis DS, Benkovics AH, Husbands AY, Timmermans M. Boundary formation through a direct threshold-based readout of mobile small RNA gradients. Dev Cell. 2017:43(3):265–273.e6. 10.1016/j.devcel.2017.10.003 [DOI] [PubMed] [Google Scholar]
- Sladek FM. What are nuclear receptor ligands? . Mol Cell Endocrinol. 2011:334(1-2):3–13. 10.1016/j.mce.2010.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smetana O, Mäkilä R, Lyu M, Amiryousefi A, Sánchez Rodríguez F, Wu MF, Solé-Gil A, Leal Gavarrón M, Siligato R, Miyashima S, et al. . High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature. 2019:565(7740):485–489. 10.1038/s41586-018-0837-0 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc. 2006:1(4):2019–2025. 10.1038/nprot.2006.286 [DOI] [PubMed] [Google Scholar]
- Stanislas T, Platre MP, Liu M, Rambaud-Lavigne L, Jaillais Y, Hamant O. A phosphoinositide map at the shoot apical meristem in Arabidopsis thaliana. BMC Biol. 2018:16(1):20. 10.1186/s12915-018-0490-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillman MC, Imai N, Li Y, Khadka M, Okafor CD, Juneja P, Adhiyaman A, Hagen SJ, Cohen DE, Ortlund EA. Allosteric regulation of thioesterase superfamily member 1 by lipid sensor domain binding fatty acids and lysophosphatidylcholine. Proc Natal Acad Sci U S A. 2020:117(36):22080–22089. 10.1073/pnas.2003877117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujishita Y, Hurley JH. Structure and lipid transport mechanism of a StAR-related domain. Nat Struct Biol. 2000:7(5):408–414. 10.1038/75192 [DOI] [PubMed] [Google Scholar]
- Van Leene J, Eeckhout D, Cannoot B, De Winne N, Persiau G, Van De Slijke E, Vercruysse L, Dedecker M, Verkest A, Vandepoele K, et al. . An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat Protoc. 2015:10(1):169–187. 10.1038/nprot.2014.199 [DOI] [PubMed] [Google Scholar]
- Wang S, Li L, Li H, Sahu SK, Wang H, Xu Y, Xian W, Song B, Liang H, Cheng S, et al. . Genomes of early-diverging streptophyte algae shed light on plant terrestrialization. Nat Plants. 2020:6(2):95–106. 10.1038/s41477-019-0560-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X. Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J Biol Chem. 2002:277(35):31994–32002. 10.1074/jbc.M205375200 [DOI] [PubMed] [Google Scholar]
- Wenkel S, Emery J, Hou BH, Evans MM, Barton MK. A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell. 2007:19(11):3379–3390. 10.1105/tpc.107.055772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, Levine TP. Lipid transfer proteins do their thing anchored at membrane contact sites… but what is their thing?. Biochem Soc Transac. 2016:44(2):517–527. 10.1042/BST20150275 [DOI] [PubMed] [Google Scholar]
- Xu Q, Li R, Weng L, Sun Y, Li M, Xiao H. Domain-specific expression of meristematic genes is defined by the LITTLE ZIPPER protein DTM in tomato. Commun Biol. 2019:2(1):134. 10.1038/s42003-019-0368-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol. 2009:16(12):1230–1236. 10.1038/nsmb.1730 [DOI] [PubMed] [Google Scholar]
- Yip HK, Floyd SK, Sakakibara K, Bowman JL. Class III HD-Zip activity coordinates leaf development in Physcomitrella patens. Dev Biol. 2016:419(1):184–197. 10.1016/j.ydbio.2016.01.012 [DOI] [PubMed] [Google Scholar]
- Zhai G, Song J, Shu T, Yan J, Jin X, He J, Yin Z. LRH-1 senses signaling from phosphatidylcholine to regulate the expansion growth of digestive organs via synergy with Wnt/β-catenin signaling in zebrafish. J Genet Genomics. 2017:44(6):307–317. 10.1016/j.jgg.2017.03.006 [DOI] [PubMed] [Google Scholar]
- Zhang L, Li X, Li D, Sun Y, Li Y, Luo Q, Liu Z, Wang J, Li X, Zhang H, et al. . CARK1 Mediates ABA signaling by phosphorylation of ABA receptors. Cell Discov. 2018:4(1): 30. 10.1038/s41421-018-0029-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article or the supporting information.