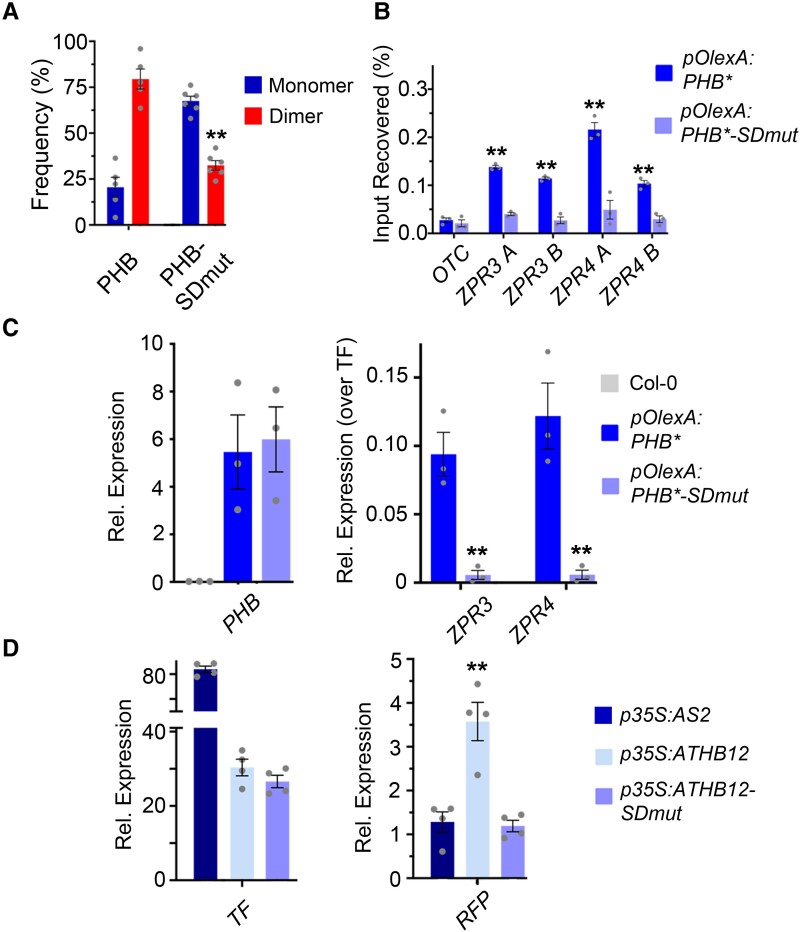
Figure 5.
Mutating PHB START reduces homodimerization and abolishes DNA-binding competence. A) Percentages of two-step photobleaching events from SiMPull translate to dimerization frequencies of ∼80% for PHB and ∼40% for PHB-SDmut. B) PHB-SDmut does not bind ZPR3 or ZPR4 regulatory regions occupied by PHB. C) Unlike PHB, PHB-SDmut cannot activate ZPR3 or ZPR4 targets in 24 h estradiol-induction experiments. D) Domain-capture-mimicry experiments demonstrate fusion of SDmut START to ATHB12 (ATHB-SDmut) renders ATHB12 non-functional, as RFP reporter levels are indistinguishable from the AS2 negative control. n = 3 biological replicates. **P ≤ 0.01, Student's t-test. Note: PHB, PHB-SDmut, and PHB-Delta data were collected simultaneously. PHB data from Figs. 2 and 3 are replotted for clarity.