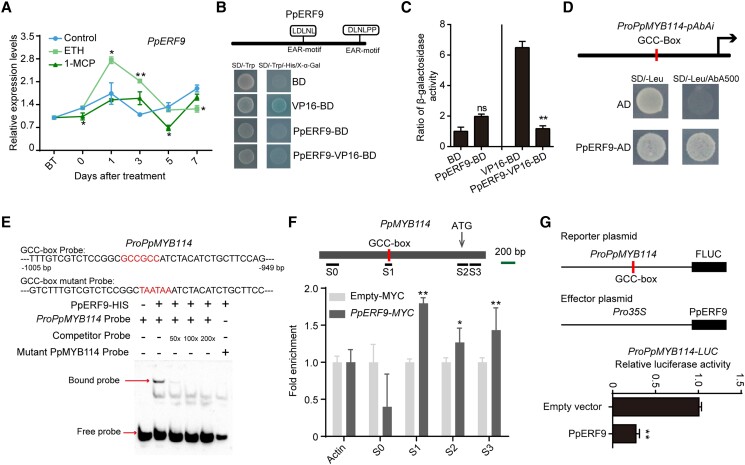
Figure 1.
Ethylene-induced PpERF9 represses PpMYB114 transcription. A) Expression pattern of PpERF9 in “Hongzaosu” pear fruit following ethephon and 1-MCP treatments. B) Transcriptional repression associated with PpERF9 in yeast cells. C) The β-galactosidase activities reflect the transcriptional repression by PpERF9. D) A Y1H assay indicated that PpERF9 can bind directly to the PpMYB114 promoter containing a GCC-box. E) An EMSA confirmed that PpERF9 binds directly to the GCC-box in the PpMYB114 promoter. The hot probe was a biotin-labeled fragment of the PpMYB114 promoter containing the GCC-box, whereas the competitor probe was an unlabeled probe (50-, 100-, and 200-fold molar excess). The mutant cold probe was the same as the labeled hot probe but with GCCGCC mutated to TAATAA. The His-tagged PpERF9 protein was purified. F) ChIP-qPCR assay results reflected the in vivo binding of PpERF9 to the PpMYB114 promoter. Cross-linked chromatin samples were extracted from PpERF9-MYC-overexpressing pear calli with 3 biological replicates and precipitated with an anti-MYC antibody. Eluted DNA was used to amplify the sequences neighboring the GCC-box by qPCR. Four regions (S0 to S3) were examined. The bar indicates the length of 200 bp. Pear calli overexpressing the MYC sequence were used as a negative control. Three biological replicates of pear calli were used in the ChIP assay. G) A dual-luciferase assay demonstrated that PpERF9 directly inhibits PpMYB114 promoter activity. The promoter of PpMYB114 was cloned into the pGreenII 0800-LUC (firefly luciferase) vector, and the full-length CDS of PpERF9 was cloned into the pGreenII 0029 62-SK vector. The empty vector of SK was used as control. The relative luciferase activity was analyzed. Error bars represent the standard deviation of 3 biological replicates. Asterisks indicate significantly different values (*P < 0.05 and **P < 0.01).