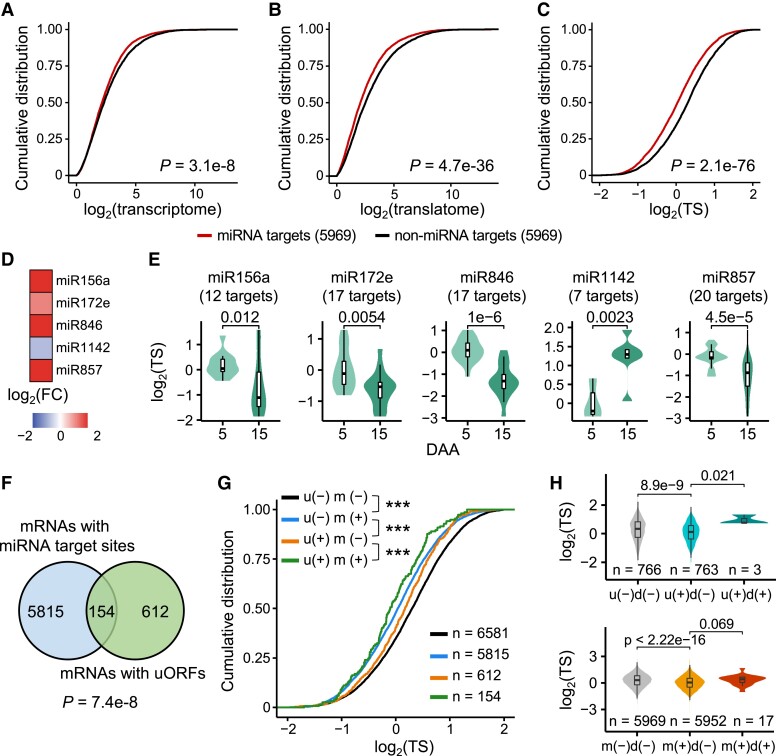
Figure 6.
Combinatorial regulation of gene expression at the translational level by upstream open reading frames (uORFs), downstream ORFs (dORFs), and microRNAs (miRNAs). A–C) Cumulative distributions of the transcriptional level (A), translational level (B), and translation state (TS) (C) of the miRNA target genes and non-miRNA target genes. The different colored lines represent mRNAs with or without miRNA target sites. Two-sided Wilcoxon tests were performed. Only expressed genes with a FPKM > 1 were retained. A random set of the gene without miRNA target sites was used as a control. D) The fold changes in expression levels of several miRNAs during grain development. The comparison of 15-DAA grains to 5-DAA grains was shown. E) The TS distributions of the targets of several miRNAs. The predicted targets with miRNA-mediated translation repression were shown. Two-sided Wilcoxon tests were performed. F) A Venn diagram illustrates the cooccurrence of miRNAs and uORFs in gene models. A hypergeometric test was performed. G) The TS distributions of the “u−m−,” “u−m+,” “u+m−,” and “u+m+” mRNAs. “u−,” genes without an uORF; “u+,” genes with an uORF; “m−,” non-miRNA target genes; “m+,” miRNA target genes. A random set of the gene without uORFs or miRNA target sites was used as a control. Two-sided Wilcoxon tests were performed. ***P < 0.001. H) dORFs enhance the TSs of mRNAs with uORFs or microRNA target sites. “d−”, genes without a dORF. “d+”, genes with a dORF. The random set of the gene without specific features was used as a control.