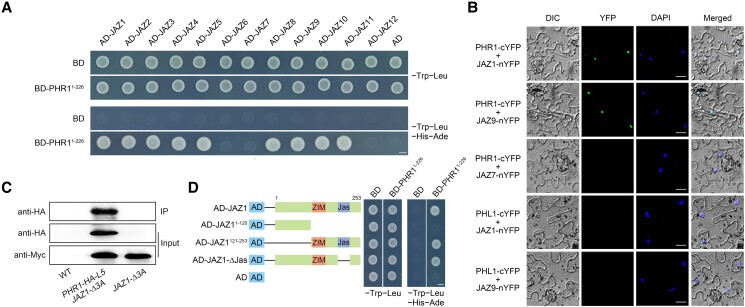
Figure 1.
Physical interactions of JAZ repressors with PHR1, PHL2, and PHL3. A) Y2H assay analyses. Protein interactions were indicated by the ability of cells to grow on dropout medium lacking Leu, Trp, His, and Ade and containing 20 mM 3-aminotriazole after a 2-d incubation. pGBKT7 (BD) and pGADT7 (AD) were used as negative controls. Bars = 2.5 mm. B) BiFC analyses. The fluorescence detected in the nucleus of transformed Nicotiana benthamiana cells co-expressing JAZ1-nYFP (or JAZ9-nYFP) with PHR1-cYFP under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Pro35S). No signal was observed in the negative controls where PHR1-cYFP and JAZ7-nYFP or PHL1-cYFP and JAZ1-nYFP (or JAZ9-nYFP) were co-expressed. Nuclei are indicated by DAPI staining. Bars = 15 µm. C) CoIP assays. Total proteins were extracted from 8-d-old transgenic Arabidopsis seedlings simultaneously overexpressing PHR1 and JAZ1 (PHR1-HA-L5 JAZ1-Δ3A) under the control of Pro35S. 3Myc-fused JAZ1 was immunoprecipitated using an anti-Myc antibody (1:250) and the co-immunoprecipitated PHR1-HA protein was detected using an anti-HA antibody (1:10,000). Protein input for 3Myc-fused JAZ1 in the immunoprecipitated complexes was also tested and is displayed. Experiments were repeated three times, with similar results. IP, immunoprecipitation. D) Y2H assay showing that the ZIM domain of JAZ1 is responsible for the interaction with PHR1. Left: diagram of the full-length and truncated JAZ1 constructs with specific deletions. Right: interactions are indicated by the ability of yeast cells to grow on the dropout medium lacking Leu, Trp, His, and Ade and containing 20 mM 3-aminotriazole after a 2-d incubation. BD and AD were used as negative controls. Bars = 2.5 mm.